3O8Z
 
 | | Crystal structure of Spn1 (Iws1) core domain | | Descriptor: | SULFATE ION, Transcription factor IWS1 | | Authors: | McDonald, S.M, Close, D, Hill, C.P. | | Deposit date: | 2010-08-03 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and biological importance of the spn1-spt6 interaction, and its regulatory role in nucleosome binding.
Mol.Cell, 40, 2010
|
|
1CZG
 
 | |
2K5G
 
 | | Solution NMR structure of protein encoded by gene BPP1335 from Bordetella parapertussis: Northeast Structural Genomics Target BpR195 | | Descriptor: | uncharacterized protein | | Authors: | Singarapu, K, Eletsky, A, Sathyamoorthy, B, Sukumaran, D, Wang, D, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein encoded by gene BPP1335 from
Bordetella parapertussis: Northeast Structural Genomics Target BpR195
To be Published
|
|
2YIQ
 
 | | Structural analysis of checkpoint kinase 2 in complex with inhibitor PV1322 | | Descriptor: | (E)-5-(1-(2-CARBAMIMIDOYLHYDRAZONO)ETHYL)-N-(1H-INDOL-6-YL)-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
1UL9
 
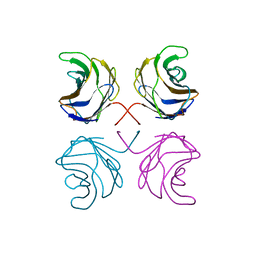 | | CGL2 ligandfree | | Descriptor: | galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
2X79
 
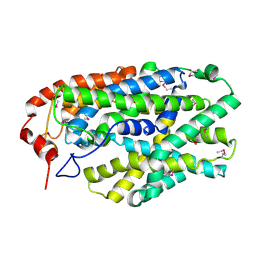 | | Inward facing conformation of Mhp1 | | Descriptor: | Hydantoin permease | | Authors: | Shimamura, T, Weyand, S, Beckstein, O, Rutherford, N.G, Hadden, J.M, Sharples, D, Sansom, M.S.P, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Basis of Alternating Access Membrane Transport by the Sodium-Hydantoin Transporter Mhp1.
Science, 328, 2010
|
|
2KPT
 
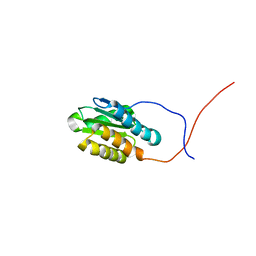 | | Solution NMR structure of the N-terminal domain of cg2496 protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR26A | | Descriptor: | Putative secreted protein | | Authors: | Eletsky, A, Sathyamoorthy, B, Sukumaran, D.K, Wang, D, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal domain of cg2496 protein from Corynebacterium glutamicum
To be Published
|
|
6V0N
 
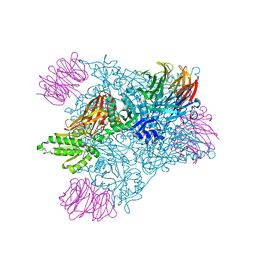 | | PRMT5 bound to PBM peptide from Riok1 | | Descriptor: | Methylosome protein 50, Protein arginine N-methyltransferase 5, Riok1 PBM peptide, ... | | Authors: | McMIllan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
2KQT
 
 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | Authors: | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
6RR7
 
 | | Influenza A virus (A/NT/60/1968) polymerase Heterotrimer bound to 3'5' vRNA promoter and capped RNA primer | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(M7G))-R(P*AP*AP*UP*CP*U)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
3FLB
 
 | |
3OAW
 
 | |
6UPJ
 
 | | HIV-2 PROTEASE/U99294 COMPLEX | | Descriptor: | 6,7,8,9-TETRAHYDRO-4-HYDROXY-3-(1-PHENYLPROPYL)CYCLOHEPTA[B]PYRAN-2-ONE, HIV-2 PROTEASE | | Authors: | Watenpaugh, K.D, Mulichak, A.M, Finzel, B.C. | | Deposit date: | 1996-12-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Use of medium-sized cycloalkyl rings to enhance secondary binding: discovery of a new class of human immunodeficiency virus (HIV) protease inhibitors.
J.Med.Chem., 38, 1995
|
|
5TZO
 
 | | Computationally Designed Fentanyl Binder - Fen49*-Complex | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase A, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
3OBH
 
 | | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104 | | Descriptor: | ACETIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Abashidze, M, Lew, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104
To be Published
|
|
4I9G
 
 | | Crystal structure of glycerol phosphate phosphatase Rv1692 from Mycobacterium tuberculosis in complex with magnesium | | Descriptor: | Glycerol 3-phosphate phosphatase, MAGNESIUM ION | | Authors: | Biswas, T, Larrouy-Maumus, G, de Carvalho, L.P, Tsodikov, O.V. | | Deposit date: | 2012-12-05 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery of a glycerol 3-phosphate phosphatase reveals glycerophospholipid polar head recycling in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1CNW
 
 | |
1CQQ
 
 | | TYPE 2 RHINOVIRUS 3C PROTEASE WITH AG7088 INHIBITOR | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, TYPE 2 RHINOVIRUS 3C PROTEASE | | Authors: | Matthews, D, Ferre, R.A. | | Deposit date: | 1999-08-10 | | Release date: | 1999-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3FPJ
 
 | | Crystal Structure of E81Q mutant of MtNAS in complex with S-ADENOSYLMETHIONINE | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, Putative uncharacterized protein, ... | | Authors: | Dreyfus, C, Pignol, D, Arnoux, P. | | Deposit date: | 2009-01-05 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic snapshots of iterative substrate translocations during nicotianamine synthesis in Archaea
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3BRD
 
 | | CSL (Lag-1) bound to DNA with Lin-12 RAM peptide, P212121 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
9BDV
 
 | | NF-kappaB RelA homo-dimer bound to TA-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6USC
 
 | | Structure of Human Intelectin-1 in complex with KO | | Descriptor: | CALCIUM ION, CHLORIDE ION, Intelectin-1, ... | | Authors: | Windsor, I.W, Isabella, C.R, Kosma, P, Raines, R.T, Kiessling, L.L. | | Deposit date: | 2019-10-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Stereoelectronic Effects Impact Glycan Recognition.
J.Am.Chem.Soc., 142, 2020
|
|
1M2B
 
 | | Crystal structure at 1.25 Angstroms resolution of the Cys55Ser variant of the thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, [2Fe-2S] ferredoxin | | Authors: | Yeh, A.P, Ambroggio, X.I, Andrade, S.L.A, Einsle, O, Chatelet, C, Meyer, J, Rees, D.C. | | Deposit date: | 2002-06-22 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution crystal structures
of the wild type and Cys-55-->Ser and
Cys-59-->Ser variants of the thioredoxin-like
[2Fe-2S] ferredoxin from Aquifex aeolicus
J.Biol.Chem., 277, 2002
|
|
4HUP
 
 | | Structure of ricin A chain bound with N-(N-(pterin-7-yl)carbonylglycyl)-L-phenylalanyl)-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-1H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-3-phenyl-propanoic acid, MALONIC ACID, Ricin, ... | | Authors: | Jasheway, K.R, Monzingo, A.F, Saito, R, Pruet, J.M, Manzano, L.A, Wiget, P.A, Anslyn, E.V, Robertus, J.D. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Peptide-conjugated pterins as inhibitors of ricin toxin A.
J.Med.Chem., 56, 2013
|
|
2L9I
 
 | |
