8ZNB
 
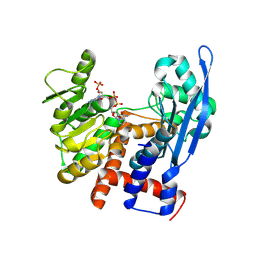 | |
8ZN4
 
 | | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution
To Be Published
|
|
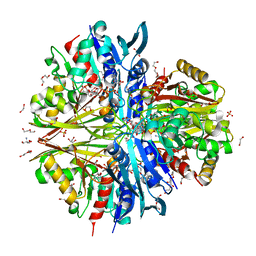
8ZN3
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution.
To Be Published
|
|
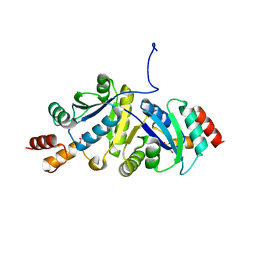
8ZN2
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution.
To Be Published
|
|
8ZMU
 
 | |
8ZMG
 
 | | Crystal structure of an inverse agonist antipsychotic drug pimavanserin-bound 5-HT2A | | Descriptor: | 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, Pimavanserin | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
8ZMF
 
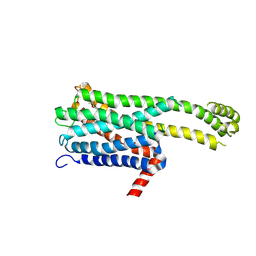 | | Crystal structure of an inverse agonist antipsychotic drug derivative-bound 5-HT2C | | Descriptor: | 1-[(4-fluorophenyl)methyl]-1-[(8~{S})-5-methyl-5-azaspiro[2.5]octan-8-yl]-3-[[4-(2-methylpropoxy)phenyl]methyl]urea, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562 | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
8ZM2
 
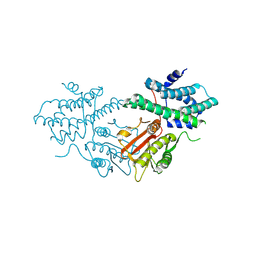 | | Structure of human pyruvate dehydrogenase kinase 2 complexed with compound 16 | | Descriptor: | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial, methyl (9~{R})-9-oxidanyl-9-(trifluoromethyl)fluorene-4-carboxylate | | Authors: | Akai, S, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-22 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design and synthesis of novel fluorene derivatives as inhibitors of pyruvate dehydrogenase kinase.
Bioorg.Med.Chem.Lett., 109, 2024
|
|
8ZM1
 
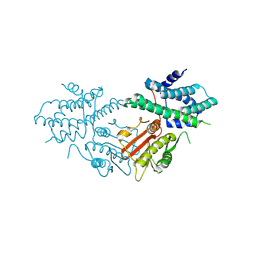 | | Structure of human pyruvate dehydrogenase kinase 2 complexed with compound 6 | | Descriptor: | (5~{R})-5-propan-2-ylindeno[1,2-b]pyridin-5-ol, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial | | Authors: | Akai, S, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-22 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Design and synthesis of novel fluorene derivatives as inhibitors of pyruvate dehydrogenase kinase.
Bioorg.Med.Chem.Lett., 109, 2024
|
|
8ZLY
 
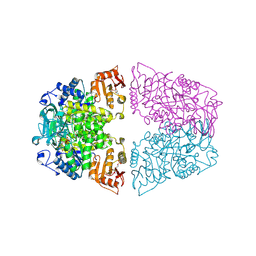 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate and UDP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2024-05-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of Nucleotide Selectivity in Pyruvate Kinase.
J.Mol.Biol., 436, 2024
|
|
8ZLE
 
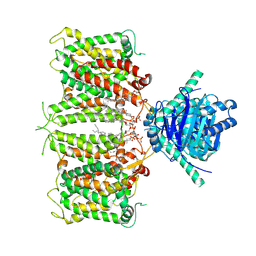 | | hAE3NTD2TMD with PT5,CLR, and Y01 | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate, ... | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-05-19 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8ZKV
 
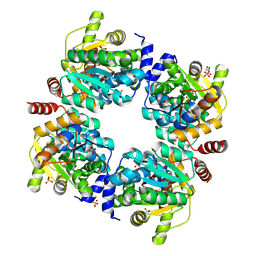 | |
8ZKM
 
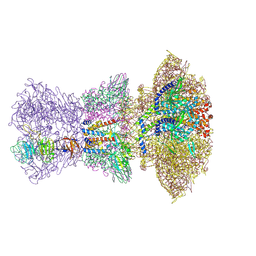 | |
8ZKK
 
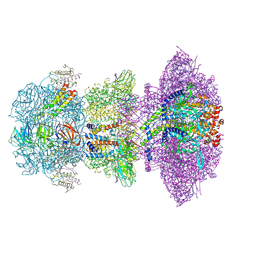 | |
8ZKD
 
 | | The Crystal Structure of the RON from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Macrophage-stimulating protein receptor beta chain, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Pan, W. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the RON from Biortus.
To Be Published
|
|
8ZKC
 
 | | iron-sulfur cluster transfer protein ApbC | | Descriptor: | GLYCEROL, Iron-sulfur cluster carrier protein, MAGNESIUM ION | | Authors: | Yang, J, Liu, L. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the iron-sulfur cluster transfer protein ApbC from Escherichia coli.
Biochem.Biophys.Res.Commun., 722, 2024
|
|
8ZJM
 
 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 5) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJL
 
 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 4) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJK
 
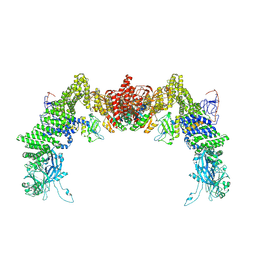 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 3) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJJ
 
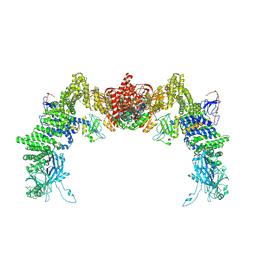 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 2) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJI
 
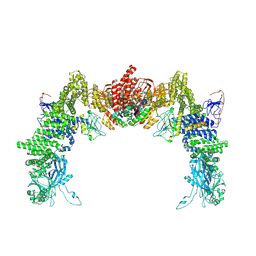 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 1) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJF
 
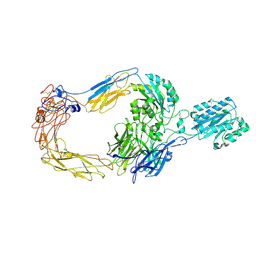 | | Cryo-EM structure of human integrin alpha-E beta-7 | | Descriptor: | CALCIUM ION, Integrin alpha-E, Integrin beta-7, ... | | Authors: | Akasaka, H, Nureki, O, Kise, Y. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of I domain-containing integrin alpha E beta 7.
Biochem.Biophys.Res.Commun., 721, 2024
|
|
8ZJ3
 
 | | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 4-Hydroxybenzoate. | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxythreonine-4-phosphate dehydrogenase, GLYCEROL, ... | | Authors: | Kumar, K.A, Pahwa, D, Kumar, P. | | Deposit date: | 2024-05-14 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 4-Hydroxybenzoate.
To Be Published
|
|
8ZJ2
 
 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZHA
 
 | | HIV-1 integrase core domain in complex with compound 15 | | Descriptor: | (2~{S})-2-[7-(cycloheptylcarbamoyl)-4',5-dimethyl-spiro[1,2-dihydroindene-3,1'-cyclohexane]-4-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase, SULFATE ION, ... | | Authors: | Furuzono, T, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and synthesis of novel and potent allosteric HIV-1 integrase inhibitors with a spirocyclic moiety.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
