1INE
 
 | |
9DKP
 
 | | The structure of human vacuolar protein sorting 34 catalytic domain bound to RD-I-53 | | Descriptor: | (8R)-3-(1,3-benzothiazol-2-yl)pyrazolo[1,5-a]pyrimidine, 2-ETHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Abiodun, W.O, Tsubaki, E, Dass, R, Singleton, J.D, Samarawickrama, P, Doukov, T, Peterson, M.A, Moody, J.D. | | Deposit date: | 2024-09-09 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Facile Bacterial Production of Human Vacuolar Protein Sorting 34 Enables Structural Characterization of Novel Inhibitors
To Be Published
|
|
5NPP
 
 | | 2.22A STRUCTURE OF THIOPHENE2 AND GSK945237 WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DIMETHYL SULFOXIDE, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Chan, P.F, Stavenger, R.A. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5JAP
 
 | |
5DJL
 
 | |
6EYZ
 
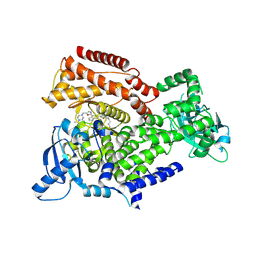 | | PI3 kinase delta in complex with 4-Fluorophenyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylic acid, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
7GXA
 
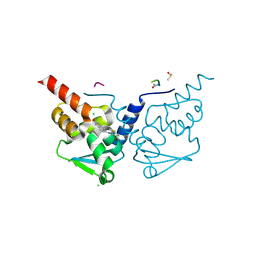 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 8 at 1.81 MGy X-ray dose. | | Descriptor: | 5-{[5-chloro-2-(methylsulfanyl)pyrimidin-4-yl]amino}-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
3EZ1
 
 | |
5JLJ
 
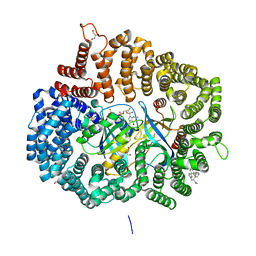 | | Crystal Structure of KPT8602 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, CHLORIDE ION, Exportin-1, ... | | Authors: | Fung, H.Y, Chook, Y.M. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies.
Leukemia, 30, 2016
|
|
5H9R
 
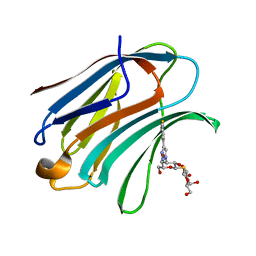 | | Crystal Structure of Human Galectin-3 CRD in Complex with TAZTDG | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
7S1G
 
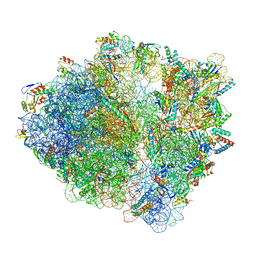 | | wild-type Escherichia coli stalled ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3ISM
 
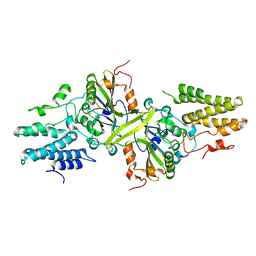 | | Crystal structure of the EndoG/EndoGI complex: Mechanism of EndoG inhibition | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CG4930, CG8862, ... | | Authors: | Loll, B, Gebhardt, M, Wahle, E, Meinhart, A. | | Deposit date: | 2009-08-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the EndoG/EndoGI complex: mechanism of EndoG inhibition.
Nucleic Acids Res., 37, 2009
|
|
3F2O
 
 | | Crystal Structure of human splA/ryanodine receptor domain and SOCS box containing 1 (SPSB1) in complex with a 20-residue VASA peptide | | Descriptor: | 20-mer peptide from ATP-dependent RNA helicase vasa, SPRY domain-containing SOCS box protein 1 | | Authors: | Filippakopoulos, P, Sharpe, T, Keates, T, Murray, J.W, Savitsky, P, Roos, A, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4.
J.Mol.Biol., 401, 2010
|
|
9MRD
 
 | |
3IAQ
 
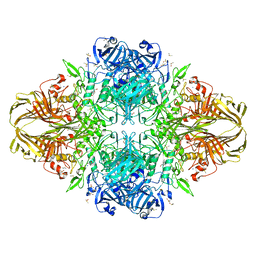 | | E. coli (lacz) beta-galactosidase (E416V) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Lo, S, Dugdale, M.L, Jeerh, N, Ku, T, Roth, N.J, Huber, R.E. | | Deposit date: | 2009-07-14 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Studies of Glu-416 variants of beta-galactosidase (E. coli) show that the active site Mg(2+) is not important for structure and indicate that the main role of Mg (2+) is to mediate optimization of active site chemistry
Protein J., 29, 2010
|
|
9MDC
 
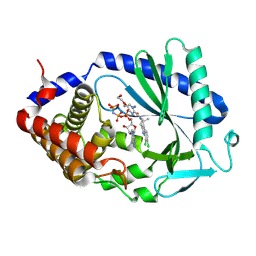 | | Crystal Structure of human cyclic GMP-AMP synthase (cGAS) in complex with compound 36; (S)-(6,7-dichloro-1-methyl-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)(5-(2-hydroxyethoxy)pyrimidin-2-yl)methanone | | Descriptor: | Cyclic GMP-AMP synthase, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sietsema, D.V. | | Deposit date: | 2024-12-05 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyrimidine Amide cGAS Inhibitors via Structure-Guided Hybridization.
Acs Med.Chem.Lett., 15, 2024
|
|
5NQ8
 
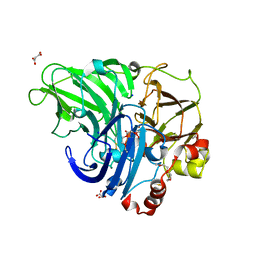 | | Crystal structure of laccases from Pycnoporus sanguineus, izoform II | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, COPPER (II) ION, Laccase, ... | | Authors: | Orlikowska, M, de J.Rostro-Alanis, M, Bujacz, A, Hernandez-Luna, C, Rubio, R, Parra, R, Bujacz, G. | | Deposit date: | 2017-04-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of two thermostable laccases from the white-rot fungus Pycnoporus sanguineus.
Int. J. Biol. Macromol., 107, 2018
|
|
7B48
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R273H mutant bound to MQ: R273H-MQ (II) | | Descriptor: | (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Degtjarik, O, Rozenberg, H, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
6LEU
 
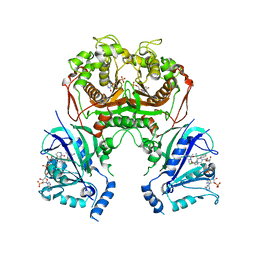 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 42 and NADPH | | Descriptor: | 1-[3-[(4-chlorophenyl)-(phenylmethyl)amino]propoxy]-6,6-dimethyl-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2019-11-27 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: Binding strengths, modes of binding and their antimalarial activities.
Eur.J.Med.Chem., 195, 2020
|
|
5T3Q
 
 | |
3BEE
 
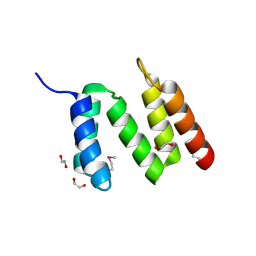 | | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Putative YfrE protein | | Authors: | Chang, C, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus.
To be Published
|
|
3IKG
 
 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-(3-methylphenyl)propyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Parge, H, Ferre, R.A, Greasley, S, Matthews, D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IEQ
 
 | |
6B31
 
 | | Structure of RORgt in complex with a novel inverse agonist 2 | | Descriptor: | (3S)-N~1~-(3-chloro-4-cyanophenyl)-N~5~-(1,3-diethyl-2,4-dioxo-1,2,3,4-tetrahydroquinazolin-6-yl)-3-methylpentanediamide, Nuclear receptor ROR-gamma | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Identification of novel quinazolinedione derivatives as ROR gamma t inverse agonist.
Bioorg. Med. Chem., 26, 2018
|
|
1CYI
 
 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
