8VRT
 
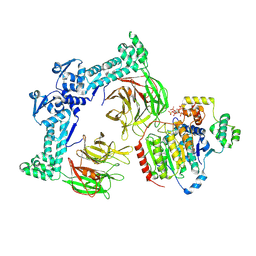 | | The structure of LSD1-CoREST-HDAC1 in complex with KBTBD4R313PRR mutant | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Xie, X, Liau, B, Zheng, N. | | Deposit date: | 2024-01-22 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations.
Nature, 639, 2025
|
|
7SU3
 
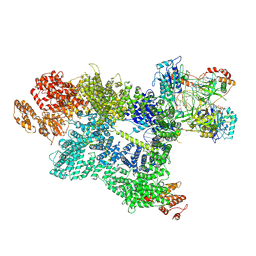 | | CryoEM structure of DNA-PK complex VII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7SGL
 
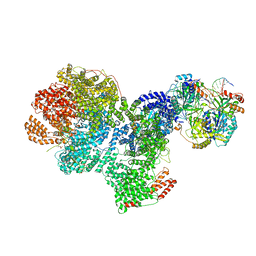 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
9E8O
 
 | |
7ZI4
 
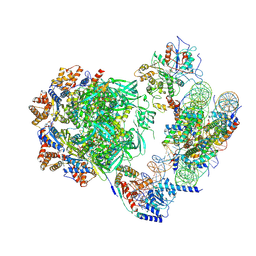 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
3WA9
 
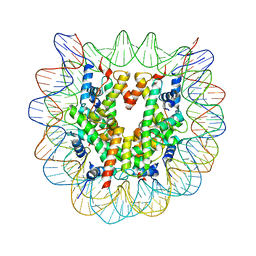 | | The nucleosome containing human H2A.Z.1 | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Sato, K, Shimada, K, Arimura, Y, Osakabe, A, Tachiwana, H, Iwasaki, W, Kagawa, W, Harata, M, Kimura, H, Kurumizaka, H. | | Deposit date: | 2013-04-30 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2HYE
 
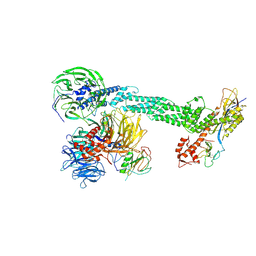 | | Crystal Structure of the DDB1-Cul4A-Rbx1-SV5V Complex | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, Nonstructural protein V, ... | | Authors: | Angers, S, Li, T, Yi, X, MacCoss, M.J, Moon, R.T, Zheng, N. | | Deposit date: | 2006-08-05 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery.
Nature, 443, 2006
|
|
7S0U
 
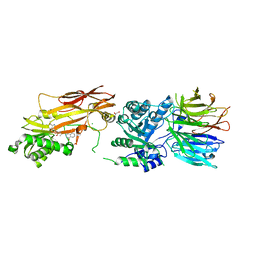 | | PRMT5/MEP50 crystal structure with MTA and phthalazinone fragment bound | | Descriptor: | 1,2-ETHANEDIOL, 4-(aminomethyl)phthalazin-1(2H)-one, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Smith, C.R, Kulyk, S, Marx, M.A. | | Deposit date: | 2021-08-31 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
7S1R
 
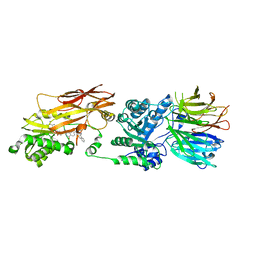 | | PRMT5/MEP50 crystal structure with MTA and a phthalazinone inhibitor bound (compound (M)-31) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, Protein arginine N-methyltransferase 5, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Kulyk, S, Smith, C.R, Marx, M.A. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
7S1S
 
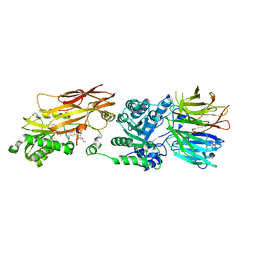 | | PRMT5/MEP50 crystal structure with MTA and MRTX-1719 bound | | Descriptor: | (7-{(5M)-5-[3-chloro-6-cyano-5-(cyclopropyloxy)-2-fluorophenyl]-1-methyl-1H-pyrazol-4-yl}-4-oxo-3,4-dihydrophthalazin-1-yl)methanaminium, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Kulyk, S, Smith, C.R, Marx, M.A. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
8B7W
 
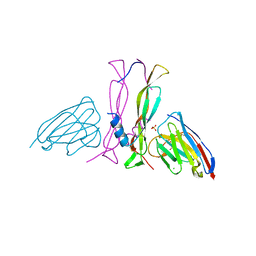 | | Complex IL-17A/anti-IL-17A-76 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-17A, ... | | Authors: | Kostareva, O.S, Svoeglazova, A, Kolyadenko, I.A, Dzhus, U.F, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2022-10-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two Epitope Regions Revealed in the Complex of IL-17A and Anti-IL-17A V H H Domain.
Int J Mol Sci, 23, 2022
|
|
4XMN
 
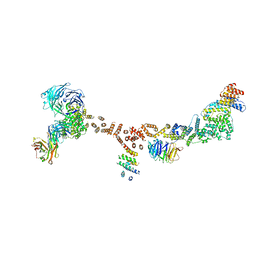 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XMM
 
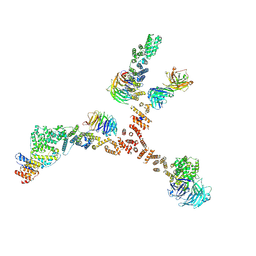 | | Structure of the yeast coat nucleoporin complex, space group C2 | | Descriptor: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (7.384 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
8USB
 
 | |
9E8N
 
 | |
8J07
 
 | |
9E8J
 
 | |
9E8I
 
 | |
9E8Q
 
 | |
9E8H
 
 | |
5A5B
 
 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7SUD
 
 | | CryoEM structure of DNA-PK complex VIII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
9E8K
 
 | |
7UD5
 
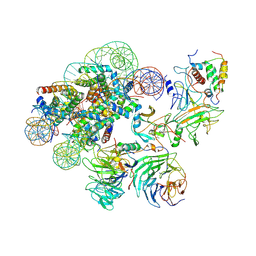 | | Complex between MLL1-WRAD and an H2B-ubiquitinated nucleosome | | Descriptor: | 601 DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Niklas, H.A, Rahman, S, Worden, E.J, Wolberger, C. | | Deposit date: | 2022-03-18 | | Release date: | 2022-09-21 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7C4J
 
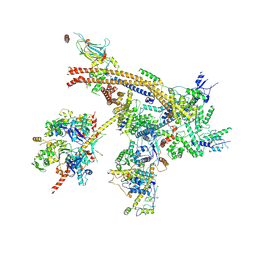 | | Cryo-EM structure of the yeast Swi/Snf complex in a nucleosome free state | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Regulator of Ty1 transposition protein 102, ... | | Authors: | Wang, C.C, Guo, Z.Y, Zhan, X.C, Zhang, X.F. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the yeast Swi/Snf complex in a nucleosome free state
Nat Commun, 2020
|
|
