1EZC
 
 | |
1EZB
 
 | |
1LXA
 
 | | UDP N-ACETYLGLUCOSAMINE ACYLTRANSFERASE | | Descriptor: | UDP N-ACETYLGLUCOSAMINE O-ACYLTRANSFERASE | | Authors: | Roderick, S.L. | | Deposit date: | 1995-10-07 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase.
Science, 270, 1995
|
|
4RF9
 
 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas in complex with L-arginine and ATPgS | | Descriptor: | ACETATE ION, ARGININE, Arginine kinase, ... | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1XK6
 
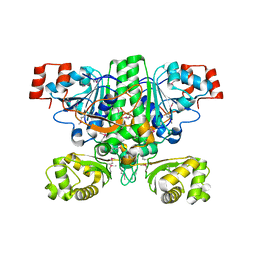 | | Crystal Structure- P1 form- of Escherichia coli Crotonobetainyl-CoA: carnitine CoA Transferase (CaiB) | | Descriptor: | Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1L9J
 
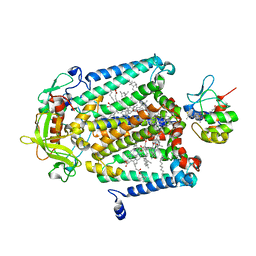 | | X-Ray Structure of the Cytochrome-c(2)-Photosynthetic Reaction Center Electron Transfer Complex from Rhodobacter sphaeroides in Type I Co-Crystals | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Okamura, M.Y, Yeh, A.P, Rees, D.C, Feher, G. | | Deposit date: | 2002-03-24 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | X-ray structure determination of the cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides.
J.Mol.Biol., 319, 2002
|
|
2E28
 
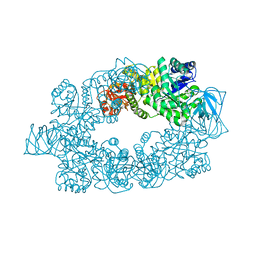 | |
1VI1
 
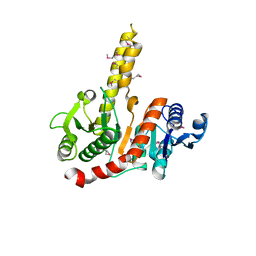 | |
4GEV
 
 | | E. coli thymidylate synthase Y209W variant in complex with substrate and a cofactor analog | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-deoxy-5'-uridylic acid, Thymidylate synthase | | Authors: | Newby, Z, Lee, T.T, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A remote mutation affects the hydride transfer by disrupting concerted protein motions in thymidylate synthase.
J.Am.Chem.Soc., 134, 2012
|
|
4GFP
 
 | | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in a second conformational state | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in second conformational state
TO BE PUBLISHED
|
|
5W8P
 
 | | Homoserine transacetylase MetX from Mycobacterium abscessus | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase, PHOSPHATE ION, ... | | Authors: | Rodriguez, E.S, Reed, R.W, Korotkov, K.V. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of mycobacterial homoserine transacetylases central to methionine biosynthesis reveals druggable active site.
Sci Rep, 9, 2019
|
|
8ODQ
 
 | | SufS-SufU complex from Mycobacterium tuberculosis | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Cysteine desulfurase, NITRATE ION, ... | | Authors: | Elchennawi, I, Carpentier, P, Caux, C, Ponge, M, Ollagnier de Choudens, S. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Characterization of Mycobacterium tuberculosis Zinc SufU-SufS Complex.
Biomolecules, 13, 2023
|
|
1Z1E
 
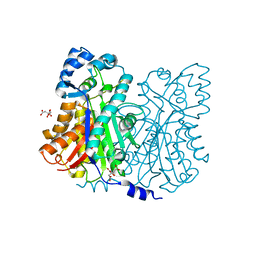 | | Crystal structure of stilbene synthase from Arachis hypogaea | | Descriptor: | CITRIC ACID, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
8K1C
 
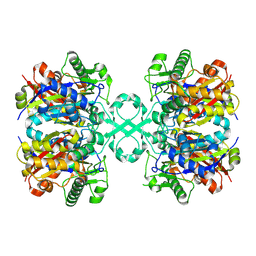 | |
1Z4E
 
 | |
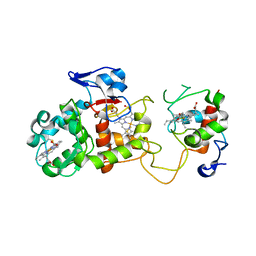
2OZ1
 
 | |
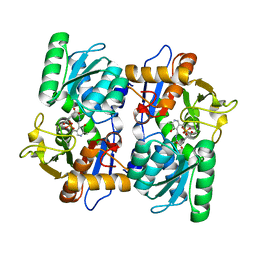
2EFY
 
 | |
3ZEI
 
 | | Structure of the Mycobacterium tuberculosis O-Acetylserine Sulfhydrylase (OASS) CysK1 in complex with a small molecule inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[(Z)-[(5Z)-5-[[2-(2-hydroxy-2-oxoethyloxy)phenyl]methylidene]-3-methyl-4-oxidanylidene-1,3-thiazolidin-2-ylidene]amino]benzoic acid, O-ACETYLSERINE SULFHYDRYLASE, ... | | Authors: | Poyraz, O, Schnell, R, Schneider, G. | | Deposit date: | 2012-12-05 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Novel Thiazolidine Inhibitors of O-Acetyl Serine Sulfhydrylase from Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
3ZBN
 
 | | Crystal structure of SCP2 thiolase from Leishmania mexicana. Complex of the C123A mutant with Coenzyme A. | | Descriptor: | 3-KETOACYL-COA THIOLASE-LIKE PROTEIN, COENZYME A | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2012-11-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
4LI3
 
 | |
1VKC
 
 | | Putative acetyl transferase from Pyrococcus furiosus | | Descriptor: | IODIDE ION, putative acetyl transferase | | Authors: | Habel, J.E, Liu, Z.-J, Tempel, W, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Putative acetyl transferase from Pyrococcus furiosus
To be published
|
|
4QUS
 
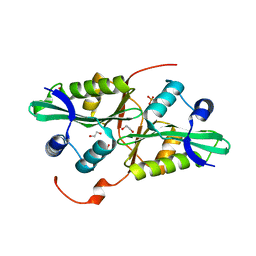 | | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase YpeA, PHOSPHATE ION | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Flores, K.J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655
To be Published
|
|
3ZBG
 
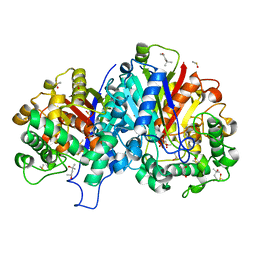 | | Crystal structure of wild-type SCP2 thiolase from Leishmania mexicana at 1.85 A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-KETOACYL-COA THIOLASE-LIKE PROTEIN, DIMETHYL SULFOXIDE | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2012-11-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
4RCR
 
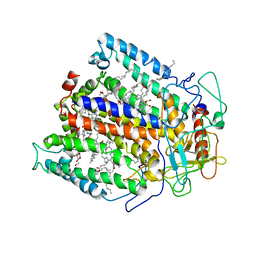 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
2E2E
 
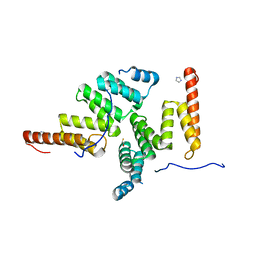 | | TPR domain of NrfG mediates the complex formation between heme lyase and formate-dependent nitrite reductase in Escherichia Coli O157:H7 | | Descriptor: | BETA-MERCAPTOETHANOL, Formate-dependent nitrite reductase complex nrfG subunit, IMIDAZOLE | | Authors: | Han, D, Kim, K, Oh, J, Park, J, Kim, Y. | | Deposit date: | 2006-11-11 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | TPR domain of NrfG mediates complex formation between heme lyase and formate-dependent nitrite reductase in Escherichia coli O157:H7.
Proteins, 70, 2008
|
|
