8SZK
 
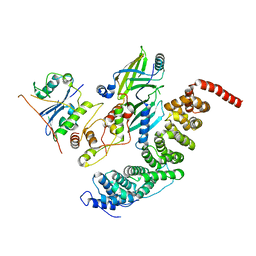 | |
8EQV
 
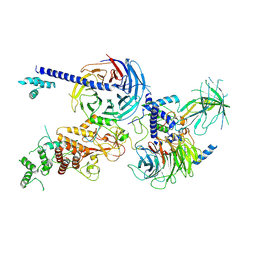 | | Cryo-EM structure of PRC2 in complex with the long isoform of AEBP2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Boudes, M, Zhang, Q, Flanigan, S.F, Davidovich, C. | | Deposit date: | 2022-10-09 | | Release date: | 2024-07-31 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | To be updated
To Be Published
|
|
7ZI4
 
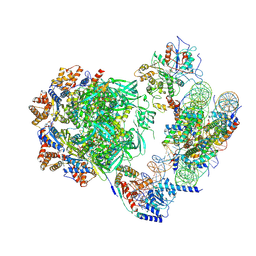 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
7SU3
 
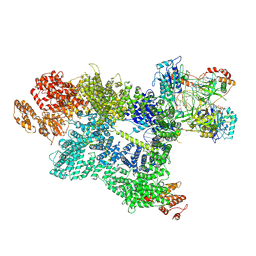 | | CryoEM structure of DNA-PK complex VII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7SGL
 
 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
1U6G
 
 | | Crystal Structure of The Cand1-Cul1-Roc1 Complex | | Descriptor: | Cullin homolog 1, RING-box protein 1, TIP120 protein, ... | | Authors: | Goldenberg, S.J, Shumway, S.D, Cascio, T.C, Garbutt, K.C, Liu, J, Xiong, Y, Zheng, N. | | Deposit date: | 2004-07-29 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases
Cell(Cambridge,Mass.), 119, 2004
|
|
8P6W
 
 | | Cryo-EM structure of CAK in complex with inhibitor BS-181 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P6Z
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0510-R | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P72
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0768 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P73
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0829 | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidine-3,4-diol, (3S,4S)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidine-3,4-diol, CDK-activating kinase assembly factor MAT1, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8PLZ
 
 | | Cryo-EM structure of CAK in complex with inhibitor CT7030 | | Descriptor: | (3~{R},4~{R})-4-[[[7-[(2-methoxyphenyl)methylamino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-06-27 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P70
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0510-S | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P6X
 
 | | Cryo-EM structure of CAK in complex with inhibitor BS-194 | | Descriptor: | (2S,3S)-3-{[7-(benzylamino)-3-(1-methylethyl)pyrazolo[1,5-a]pyrimidin-5-yl]amino}butane-1,2,4-triol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P74
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0880 (ring-up conformation) | | Descriptor: | (2S,3S)-3-[[7-[(2-bromophenyl)methylamino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]butane-1,2,4-triol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P75
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0880 (ring-down conformation) | | Descriptor: | (2S,3S)-3-[[7-[(2-bromophenyl)methylamino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]butane-1,2,4-triol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P76
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0914 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P71
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0574 | | Descriptor: | (3R,4S)-4-[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]pyrrolidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P77
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0943 | | Descriptor: | (3S,4S)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P78
 
 | | Cryo-EM structure of CAK in complex with inhibitor dinaciclib | | Descriptor: | 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P79
 
 | | Cryo-EM structure of CAK with averaged inhibitor density | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7 | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
9D8P
 
 | | Focused map of Cryo-EM structure of Ubiquitin C-degron bound to KLHDC10-EloB/C | | Descriptor: | Elongin-B, Elongin-C, Kelch domain-containing protein 10, ... | | Authors: | Chittori, S, Scott, D.C, Schulman, B.A. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-20 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for C-degron selectivity across KLHDCX family E3 ubiquitin ligases.
Nat Commun, 15, 2024
|
|
9DDE
 
 | | ncPRC1RYBP bound to H2AK119Ub/H1.4 chromatosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H1.4, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-28 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
8VAT
 
 | |
7SUD
 
 | | CryoEM structure of DNA-PK complex VIII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
3DZU
 
 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with BVT.13, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2-[(2,4-DICHLOROBENZOYL)AMINO]-5-(PYRIMIDIN-2-YLOXY)BENZOIC ACID, DNA (5'-D(*DCP*DAP*DAP*DAP*DCP*DTP*DAP*DGP*DGP*DTP*DCP*DAP*DAP*DAP*DGP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
