1BTL
 
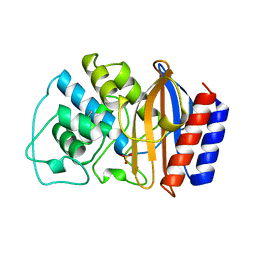 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI TEM1 BETA-LACTAMASE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | BETA-LACTAMASE TEM1, SULFATE ION | | Authors: | Jelsch, C, Mourey, L, Masson, J.M, Samama, J.P. | | Deposit date: | 1993-11-01 | | Release date: | 1995-01-26 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution.
Proteins, 16, 1993
|
|
2VUR
 
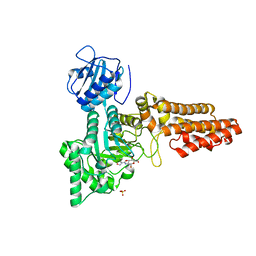 | | Chemical dissection of the link between Streptozotocin, O-GlcNAc and pancreatic cell death | | Descriptor: | 2-deoxy-2-{[(2-hydroxy-1-methylhydrazino)carbonyl]amino}-beta-D-glucopyranose, O-GLCNACASE NAGJ, SULFATE ION | | Authors: | Pathak, S, Dorfmueller, H.C, Borodkin, V.S, van Aalten, D.M.F. | | Deposit date: | 2008-05-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical Dissection of the Link between Streptozotocin, O-Glcnac, and Pancreatic Cell Death.
Chem.Biol., 15, 2008
|
|
8X43
 
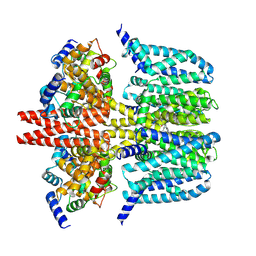 | | human KCNQ2-CaM-Ebio1-S1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(4-azanyl-1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-11-15 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 2024
|
|
8WUC
 
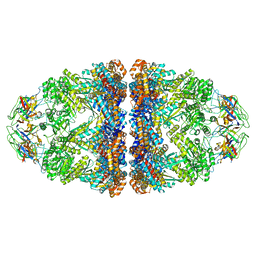 | | Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WUW
 
 | | Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WUX
 
 | | Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WU4
 
 | | Cryo-EM structure of native H. thermoluteolus TH-1 GroEL | | Descriptor: | Chaperonin GroEL | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
3MSH
 
 | |
3S9M
 
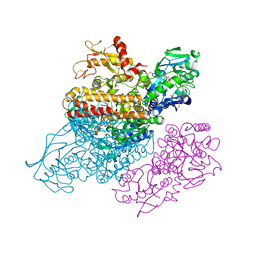 | | Complex between transferrin receptor 1 and transferrin with iron in the N-Lobe, cryocooled 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eckenroth, B.E, Steere, A.N, Mason, A.B, Everse, S.J. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S9N
 
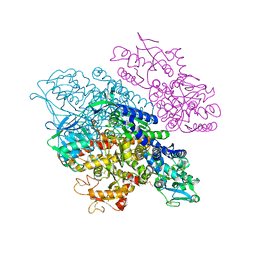 | | Complex between transferrin receptor 1 and transferrin with iron in the N-Lobe, room temperature | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eckenroth, B.E, Steere, A.N, Mason, A.B, Everse, S.J. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8V38
 
 | |
4RHW
 
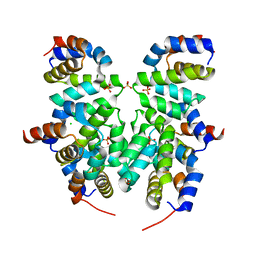 | | Crystal structure of Apaf-1 CARD and caspase-9 CARD complex | | Descriptor: | Apoptotic protease-activating factor 1, CHLORIDE ION, Caspase-9, ... | | Authors: | Hu, Q, Wu, D, Yan, C, Shi, Y. | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
7NUY
 
 | |
4Q3O
 
 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
6G72
 
 | | Mouse mitochondrial complex I in the deactive state | | Descriptor: | ACETYL GROUP, ADENOSINE-5'-DIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Agip, A.N.A, Blaza, J.N, Bridges, H.R, Viscomi, C, Rawson, S, Muench, S.P, Hirst, J. | | Deposit date: | 2018-04-04 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7OBK
 
 | |
7OBL
 
 | |
4HZI
 
 | |
4IZ6
 
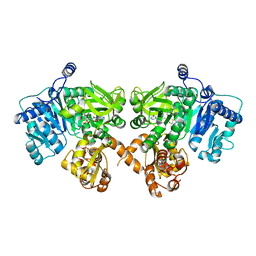 | | Structure of EntE and EntB, an NRPS adenylation-PCP fusion protein with pseudo translational symmetry | | Descriptor: | 4'-PHOSPHOPANTETHEINE, 5'-deoxy-5'-({[2-(2,3-dihydroxyphenyl)ethyl]sulfonyl}amino)adenosine, Enterobactin synthase component E, ... | | Authors: | Gulick, A.M, Sundlov, J.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure determination of the functional domain interaction of a chimeric nonribosomal peptide synthetase from a challenging crystal with noncrystallographic translational symmetry.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8OX1
 
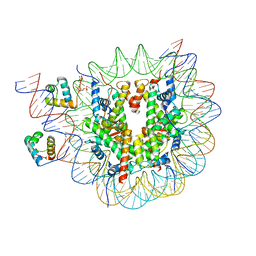 | | Structure of TRF1core in complex with telomeric nucleosome | | Descriptor: | Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, Histone H3.1, ... | | Authors: | Hu, H, van Roon, A.M.M, Ghanim, G.E, Ahsan, B, Oluwole, A, Peak-Chew, S, Robinson, C.V, Nguyen, T.H.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of telomeric nucleosome recognition by shelterin factor TRF1.
Sci Adv, 9, 2023
|
|
6G2J
 
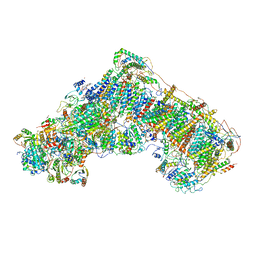 | | Mouse mitochondrial complex I in the active state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Agip, A.N.A, Blaza, J.N, Bridges, H.R, Viscomi, C, Rawson, S, Muench, S.P, Hirst, J. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-06 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1JBJ
 
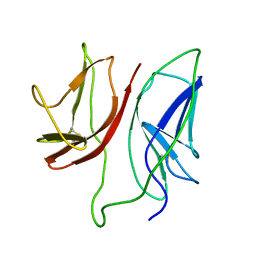 | | CD3 Epsilon and gamma Ectodomain Fragment Complex in Single-Chain Construct | | Descriptor: | CD3 Epsilon and gamma Ectodomain Fragment Complex | | Authors: | Sun, Z.-Y.J, Kim, K.S, Wagner, G, Reinherz, E.L. | | Deposit date: | 2001-06-05 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer.
Cell(Cambridge,Mass.), 105, 2001
|
|
5V03
 
 | |
5TP6
 
 | |
5TP5
 
 | |
