8E3C
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E3B
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E3A
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E39
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E38
 
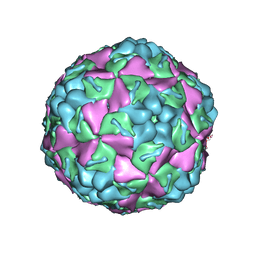 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E36
 
 | | E. coli 50S ribosome bound to compound streptogramin A analog 3146 | | Descriptor: | 2-[(3R,4R,5E,10E,12E,14S,16R,23S,26aR)-16-fluoro-14-hydroxy-12-methyl-1,7,22-trioxo-3-(propan-2-yl)-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-4-yl]-N-{2-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]ethyl}acetamide, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E35
 
 | | E. coli 50S ribosome bound to compound SAB002 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E33
 
 | | E. coli 50S ribosome bound to compound streptogramin analog SAB001 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E32
 
 | | E. coli 50S ribosome bound to compound streptogramin analogs SA1 and SB1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,23S,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-yn-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E31
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | Genome polyprotein, VP1, VP3 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E30
 
 | | E. coli 50S ribosome bound to compound streptogramin A analog 3142 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (1.91 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E2Y
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | Genome polyprotein, VP1, VP2 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E2X
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E2U
 
 | | HMPV F monomer bound to RSV-199 Fab | | Descriptor: | Fusion glycoprotein F0, RSV-199 heavy chain, RSV-199 light chain | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
8E2R
 
 | | Crystal structure of TadAC-1.14 | | Descriptor: | GLYCEROL, ZINC ION, tRNA-specific adenosine deaminase 1.14 | | Authors: | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | Deposit date: | 2022-08-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
8E2Q
 
 | | Crystal structure of TadAC-1.17 in a complex with ssDNA | | Descriptor: | DNA (5'-D(P*GP*CP*GP*GP*CP*TP*(D8A)P*CP*GP*GP*A)-3'), GLYCEROL, ZINC ION, ... | | Authors: | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | Deposit date: | 2022-08-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
8E2O
 
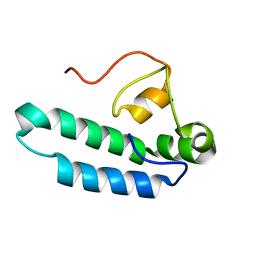 | |
8E2N
 
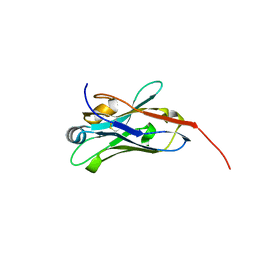 | |
8E2M
 
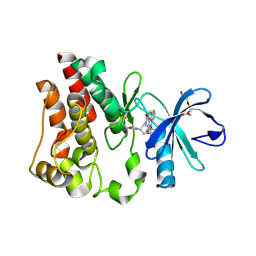 | | Bruton's tyrosine kinase (BTK) with compound 13 | | Descriptor: | (5P)-5-[4-methyl-6-(2-methylpropyl)pyridin-3-yl]-4-oxo-N-[(1R,2S)-2-propanamidocyclopentyl]-4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide, TRIETHYLENE GLYCOL, Tyrosine-protein kinase BTK | | Authors: | Alexander, R, Milligan, C.M. | | Deposit date: | 2022-08-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Discovery of JNJ-64264681: A Potent and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 65, 2022
|
|
8E2B
 
 | | N-terminal domain of S. aureus GpsB | | Descriptor: | Cell cycle protein GpsB, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylococcus aureus FtsZ and PBP4 bind to the conformationally dynamic N-terminal domain of GpsB.
Elife, 13, 2024
|
|
8E26
 
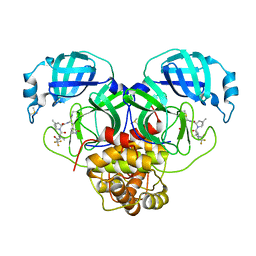 | | Crystal Structure of SARS-CoV-2 Main Protease N142S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-08-14 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8E25
 
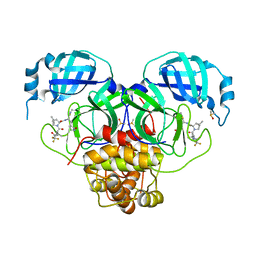 | | Crystal Structure of SARS-CoV-2 Main Protease M49I mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-08-14 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8E1Y
 
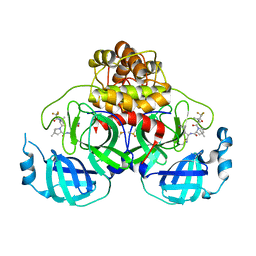 | | Crystal Structure of SARS-CoV-2 Main protease A193S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8E1X
 
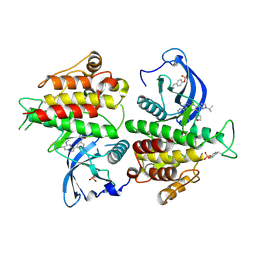 | | FGFR2 kinase domain in complex with a Pyrazolo[1,5-a]pyrimidine analog (Compound 29) | | Descriptor: | (5M)-N-methyl-5-{(6M,8S)-5-{[(3S)-oxolan-3-yl]amino}-6-[1-(propan-2-yl)-1H-pyrazol-3-yl]pyrazolo[1,5-a]pyrimidin-3-yl}pyridine-3-carboxamide, Fibroblast growth factor receptor 2 | | Authors: | Lei, H.-T, Epling, L.B, Deller, M.C. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Wild-Type and Gatekeeper Mutant Fibroblast Growth Factor Receptor (FGFR) 2/3.
J.Med.Chem., 65, 2022
|
|
8E1W
 
 | | Neutron crystal structure of Panus similis AA9A at room temperature | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Meilleur, F, Tandrup, T, Lo Leggio, L. | | Deposit date: | 2022-08-11 | | Release date: | 2023-01-11 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.1 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/neutron structure of Lentinus similis AA9_A at room temperature.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
