5CXZ
 
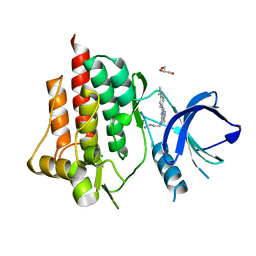 | | SYK catalytic domain complexed with naphthyridine inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, N-{7-[4-(dimethylamino)phenyl]-1,6-naphthyridin-5-yl}propane-1,3-diamine, ... | | Authors: | Thoma, G, Veenstra, S, Strang, R, Blanz, J, Vangrevelinghe, E, Berghausen, J, Lee, C.C, Zerwes, H.-G. | | Deposit date: | 2015-07-29 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Orally bioavailable Syk inhibitors with activity in a rat PK/PD model.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5V5B
 
 | | KVQIINKKLD, Structure of the amyloid spine from microtubule associated protein tau Repeat 2 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Seidler, P.M, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S, Cascio, D, Boyer, D.R. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Structure-based inhibitors of tau aggregation.
Nat Chem, 10, 2018
|
|
5D5X
 
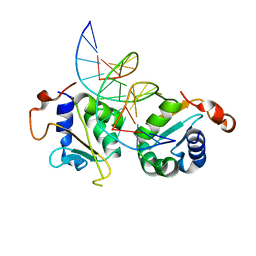 | | Crystal structure of Chaetomium thermophilum Skn7 with SSRE DNA | | Descriptor: | Putative transcription factor, SSRE DNA strand 1, SSRE DNA strand 2 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5V
 
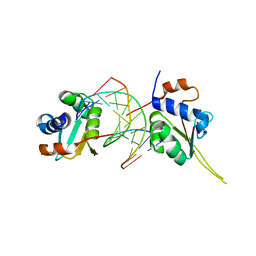 | | Crystal structure of human Hsf1 with Satellite III repeat DNA | | Descriptor: | DNA, Heat shock factor protein 1, MAGNESIUM ION | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4ZZV
 
 | | Geotrichum candidum Cel7A apo structure at 1.4A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE I, ... | | Authors: | Borisova, A.S, Stahlberg, J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Sequencing, Biochemical Characterization, Crystal Structure and Molecular Dynamics of Cellobiohydrolase Cel7A from Geotrichum Candidum 3C.
FEBS J., 282, 2015
|
|
5A8Z
 
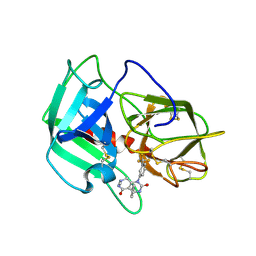 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(4R)-7-methyl-2,5-bis(oxidanylidene)-1-[3-(trifluoromethyl)phenyl]-3,4,6,8-tetrahydropyrimido[4,5-d]pyridazin-4-yl]benzenecarbonitrile, Neutrophil elastase, ... | | Authors: | von Nussbaum, F, Li, V.M, Meibom, D, Anlauf, S, Bechem, M, Delbeck, M, Gerisch, M, Harrenga, A, Karthaus, D, Lang, D, Lustig, K, Mittendorf, J, Schaefer, M, Schaefer, S, Schamberger, J. | | Deposit date: | 2015-07-17 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective Human Neutrophil Elastase Inhibitors with Novel Equatorial Ring Topology: in vivo Efficacy of the Polar Pyrimidopyridazine BAY-8040 in a Pulmonary Arterial Hypertension Rat Model.
ChemMedChem, 11, 2016
|
|
7PWF
 
 | | Cryo-EM structure of small subunit of Giardia lamblia ribosome at 2.9 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-25 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
5A7R
 
 | | Human poly(ADP-ribose) glycohydrolase in complex with synthetic dimeric ADP-ribose | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Lambrecht, M.J, Brichacek, M, Barkauskaite, E, Ariza, A, Ahel, I, Hergenrother, P.J. | | Deposit date: | 2015-07-09 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis of Dimeric Adp-Ribose and its Structure with Human Poly(Adp-Ribose) Glycohydrolase.
J.Am.Chem.Soc., 137, 2015
|
|
5D5W
 
 | | Crystal structure of Chaetomium thermophilum Skn7 with HSE DNA | | Descriptor: | HSE DNA, Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5Z
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form II | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Bracher, A, Hartl, F.U. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5DBN
 
 | | Crystal structure of AtoDA complex | | Descriptor: | Acetate CoA-transferase subunit alpha, Acetate CoA-transferase subunit beta, CHLORIDE ION, ... | | Authors: | Arbing, M.A, Koo, C.W, Shin, A, Medrano-Soto, A, Eisenberg, D. | | Deposit date: | 2015-08-21 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Crystal structure of AtoDA complex
To Be Published
|
|
7R5Z
 
 | |
6WPI
 
 | | Crystal structure of Nop9 in complex with ITS1 RNA | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, GLYCEROL, Nucleolar protein 9, ... | | Authors: | Zhang, J, Qiu, C, Hall, T.M.T. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Nop9 recognizes structured and single-stranded RNA elements of preribosomal RNA.
Rna, 26, 2020
|
|
5D5Y
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form I | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D2A
 
 | | Bifunctional dendrimers | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Michaud, G, Visini, R, Stocker, A, Darbre, T, Reymond, J.L. | | Deposit date: | 2015-08-05 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Overcoming antibiotic resistance inPseudomonas aeruginosabiofilms using glycopeptide dendrimers.
Chem Sci, 7, 2016
|
|
5D5U
 
 | | Crystal structure of human Hsf1 with HSE DNA | | Descriptor: | Heat shock Element DNA, Heat shock factor protein 1 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D60
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form III | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5DEA
 
 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA, cesium bound form. | | Descriptor: | CESIUM ION, Fragile X mental retardation protein 1, POTASSIUM ION, ... | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7973 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DAD
 
 | | Crystal Structure of Human KEAP1 BTB Domain in Complex with Small Molecule TX64014 | | Descriptor: | (6aS,7S,10aS)-8-hydroxy-4-methoxy-2,7,10a-trimethyl-5,6,6a,7,10,10a-hexahydrobenzo[h]quinazoline-9-carbonitrile, Kelch-like ECH-associated protein 1 | | Authors: | Huerta, C, Jiang, X, Trevino, I, Bender, C.F, Swinger, K.K, Stoll, V.S, Ferguson, D.A, Thomas, P.J, Probst, B, Dulubova, I, Visnick, M, Wigley, W.C. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding.
Biochim.Biophys.Acta, 1860, 2016
|
|
5DE8
 
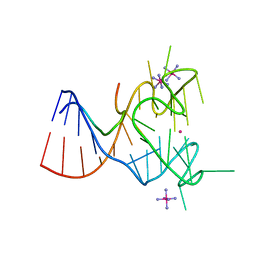 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA, iridium hexammine bound form. | | Descriptor: | Fragile X mental retardation protein 1, IRIDIUM HEXAMMINE ION, POTASSIUM ION, ... | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1003 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7R0H
 
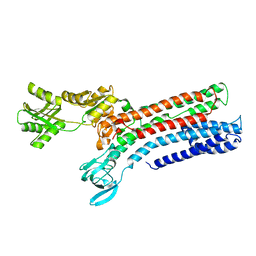 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | Descriptor: | COPPER (II) ION, Putative copper-exporting P-type ATPase A | | Authors: | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
7R0I
 
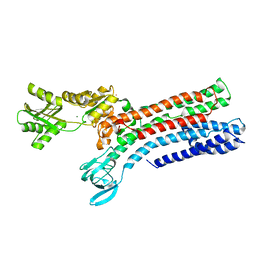 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Putative copper-exporting P-type ATPase A | | Authors: | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
7R0G
 
 | |
7RK5
 
 | |
7RK4
 
 | | Mannitol-2-dehydrogenase from Aspergillus fumigatus | | Descriptor: | Mannitol 2-dehydrogenase | | Authors: | Nguyen, S, Bruning, J.B. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting the Mannitol Biosynthesis Pathway in Aspergillus fumigatus: Characterisation and Inhibition of Mannitol-2-Dehydrogenase
To Be Published
|
|
