1PO1
 
 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R80633, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE BUTYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1YEH
 
 | | STRUCTURE OF IGG2A FAB FRAGMENT | | Descriptor: | FAB FRAGMENT, ZINC ION | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 1997-05-29 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | X-ray structures of a hydrolytic antibody and of complexes elucidate catalytic pathway from substrate binding and transition state stabilization through water attack and product release.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AFE
 
 | | HUMAN ALPHA-THROMBIN INHIBITION BY CBZ-PRO-AZALYS-ONP | | Descriptor: | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
1BJH
 
 | | HAIRPIN LOOPS CONSISTING OF SINGLE ADENINE RESIDUES CLOSED BY SHEARED A(DOT)A AND G(DOT)G PAIRS FORMED BY THE DNA TRIPLETS AAA AND GAG: SOLUTION STRUCTURE OF THE D(GTACAAAGTAC) HAIRPIN, NMR, 16 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*AP*AP*AP*GP*TP*AP*C)-3') | | Authors: | Chou, S.-H, Zhu, L, Gao, Z, Cheng, J.-W, Reid, B.R. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Hairpin loops consisting of single adenine residues closed by sheared A.A and G.G pairs formed by the DNA triplets AAA and GAG: solution structure of the d(GTACAAAGTAC) hairpin.
J.Mol.Biol., 264, 1996
|
|
1AE8
 
 | | HUMAN ALPHA-THROMBIN INHIBITION BY EOC-D-PHE-PRO-AZALYS-ONP | | Descriptor: | 1-ETHOXYCARBONYL-D-PHE-PRO-2(4-AMINOBUTYL)HYDRAZINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
1EFV
 
 | |
1JLX
 
 | | AGGLUTININ IN COMPLEX WITH T-DISACCHARIDE | | Descriptor: | AGGLUTININ, FORMYL GROUP, TOLUENE, ... | | Authors: | Transue, T.R, Smith, A.K, Mo, H, Goldstein, I.J, Saper, M.A. | | Deposit date: | 1997-07-23 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of benzyl T-antigen disaccharide bound to Amaranthus caudatus agglutinin.
Nat.Struct.Biol., 4, 1997
|
|
1AR7
 
 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT P1095S + H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1YEG
 
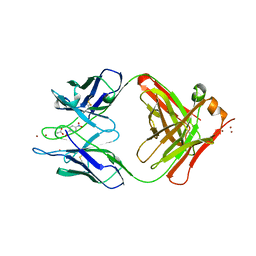 | | STRUCTURE OF IGG2A FAB FRAGMENT (D2.3) COMPLEXED WITH REACTION PRODUCT | | Descriptor: | ACETATE ION, IGG2A FAB FRAGMENT, PARANITROBENZYL ALCOHOL, ... | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 1997-05-29 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of a hydrolytic antibody and of complexes elucidate catalytic pathway from substrate binding and transition state stabilization through water attack and product release.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AR8
 
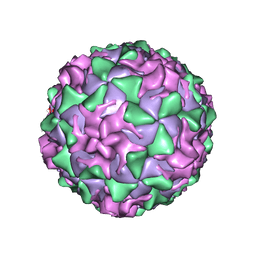 | | P1/MAHONEY POLIOVIRUS, MUTANT P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1ASJ
 
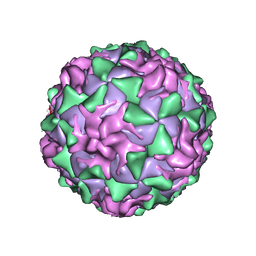 | | P1/MAHONEY POLIOVIRUS, AT CRYOGENIC TEMPERATURE | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR6
 
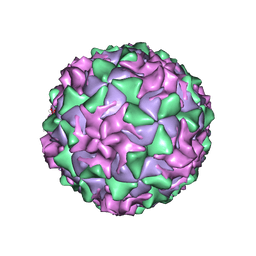 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT V1160I +P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AUI
 
 | | HUMAN CALCINEURIN HETERODIMER | | Descriptor: | CALCIUM ION, FE (III) ION, SERINE/THREONINE PHOSPHATASE 2B, ... | | Authors: | Kissinger, C.R, Parge, H.E, Knighton, D.R, Pelletier, L.A, Lewis, C.T, Tempczyk, A, Villafranca, J.E. | | Deposit date: | 1997-08-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex.
Nature, 378, 1995
|
|
1YEF
 
 | | STRUCTURE OF IGG2A FAB FRAGMENT (D2.3) COMPLEXED WITH SUBSTRATE ANALOGUE | | Descriptor: | IGG2A FAB FRAGMENT, PARA-NITROBENZYL GLUTARYL GLYCINIC ACID, ZINC ION | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 1997-05-29 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of a hydrolytic antibody and of complexes elucidate catalytic pathway from substrate binding and transition state stabilization through water attack and product release.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1ITF
 
 | | INTERFERON ALPHA-2A, NMR, 24 STRUCTURES | | Descriptor: | INTERFERON ALPHA-2A | | Authors: | Klaus, W, Gsell, B, Labhardt, A.M, Wipf, B, Senn, H. | | Deposit date: | 1997-08-22 | | Release date: | 1997-12-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution.
J.Mol.Biol., 274, 1997
|
|
1PO2
 
 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R77975, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE ETHYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1JLY
 
 | | CRYSTAL STRUCTURE OF AMARANTHUS CAUDATUS AGGLUTININ | | Descriptor: | AGGLUTININ | | Authors: | Transue, T.R, Smith, A.K, Mo, H, Goldstein, I.J, Saper, M.A. | | Deposit date: | 1997-07-23 | | Release date: | 1997-12-03 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of benzyl T-antigen disaccharide bound to Amaranthus caudatus agglutinin.
Nat.Struct.Biol., 4, 1997
|
|
1JLD
 
 | | Potent hiv protease inhibitors containing a novel (hydroxyethyl)amide isostere | | Descriptor: | (2S)-2-tert-butyl-N~4~-(1-ethylpropyl)-N~1~-[(2R,3S)-2-hydroxy-4-phenyl-3-{[N-(quinolin-2-ylcarbonyl)-L-threonyl]amino}butyl]butanediamide, Pol polyprotein | | Authors: | Tong, L. | | Deposit date: | 1997-05-31 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potent HIV protease inhibitors containing a novel (hydroxyethyl)amide isostere.
J.Med.Chem., 40, 1997
|
|
3NUL
 
 | | Profilin I from Arabidopsis thaliana | | Descriptor: | GLYCEROL, PROFILIN I, SULFATE ION | | Authors: | Thorn, K, Christensen, H.E.M, Shigeta, R, Huddler, D, Chua, N.-H, Shalaby, L, Lindberg, U, Schutt, C.E. | | Deposit date: | 1996-11-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a major allergen from plants.
Structure, 5, 1997
|
|
1MJZ
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D97N | | Descriptor: | INORGANIC PYROPHOSPHATASE | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
2EZH
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZI
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, 30 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
1JOI
 
 | | STRUCTURE OF PSEUDOMONAS FLUORESCENS HOLO AZURIN | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Lee, X, Ton-that, H, Zhu, D.-W, Biesterfedlt, J, Lanthier, P.H, Yachuchi, M, Dahms, T, Szabo, A.G. | | Deposit date: | 1997-06-09 | | Release date: | 1997-12-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallization and preliminary crystallographic studies of the crystals of the azurin Pseudomonas fluorescens.
Arch.Biochem.Biophys., 308, 1994
|
|
1FSP
 
 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, 20 STRUCTURES | | Descriptor: | STAGE 0 SPORULATION PROTEIN F | | Authors: | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | Deposit date: | 1997-06-05 | | Release date: | 1997-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
1AHN
 
 | | E. COLI FLAVODOXIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Ludwig, M.L. | | Deposit date: | 1997-04-07 | | Release date: | 1997-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A flavodoxin that is required for enzyme activation: the structure of oxidized flavodoxin from Escherichia coli at 1.8 A resolution.
Protein Sci., 6, 1997
|
|
