1SWJ
 
 | | CORE-STREPTAVIDIN MUTANT W79F AT PH 4.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWO
 
 | | CORE-STREPTAVIDIN MUTANT W120F AT PH 7.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWG
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-07-12 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
1SWF
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 | | Descriptor: | CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-04-23 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
1U06
 
 | | crystal structure of chicken alpha-spectrin SH3 domain | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Chevelkov, V, Faelber, K, Diehl, A, Heinemann, U, Oschkinat, H, Reif, B. | | Deposit date: | 2004-07-13 | | Release date: | 2005-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detection of dynamic water molecules in a microcrystalline sample of the SH3 domain of alpha-spectrin by MAS solid-state NMR.
J.Biomol.Nmr, 31, 2005
|
|
2LTH
 
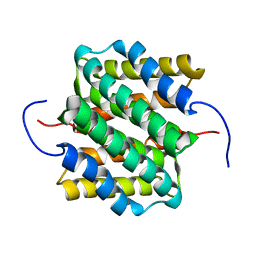 | | NMR structure of major ampullate spidroin 1 N-terminal domain at pH 5.5 | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Otikovs, M, Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S, Johansson, J. | | Deposit date: | 2012-05-25 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation.
Nat Commun, 5, 2014
|
|
2MV0
 
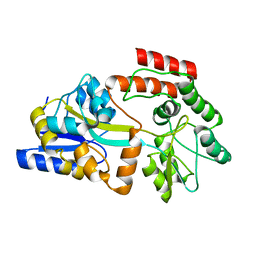 | | Solution NMR Structure of Maltose-binding protein from Escherichia coli, Northeast Structural Genomics Consortium (NESG) Target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Rossi, P, Lange, O.F, Sgourakis, N.G, Song, Y, Lee, H, Aramini, J.M, Ertekin, A, Xiao, R, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3O21
 
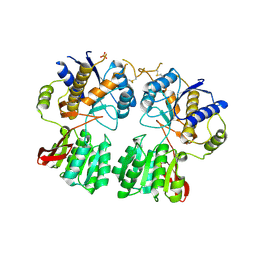 | | High resolution structure of GluA3 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 3, PHOSPHATE ION | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Babu, M.M, Jensen, M.H, Greger, I.H. | | Deposit date: | 2010-07-22 | | Release date: | 2011-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Dynamics and allosteric potential of the AMPA receptor N-terminal domain
Embo J., 30, 2011
|
|
7JR6
 
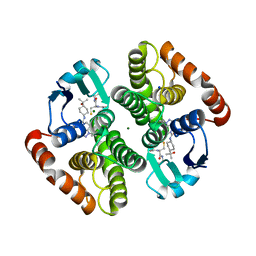 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1-(3-fluorophenyl)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7JR8
 
 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
6ZW8
 
 | | Isopenicillin N synthase in complex with Cd and ACV. | | Descriptor: | CADMIUM ION, GLYCEROL, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
8QTO
 
 | | CRYSTAL STRUCTURE OF HOLO-L28H-FNR OF A. FISCHERI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FNR type regulator, IRON/SULFUR CLUSTER | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Reactivity of [4Fe-4S] Fumarate and Nitrate Reduction (FNR) Regulator with O2 and NO: Increased O2 Resistance and Relative Specificity for NO of the [4Fe-4S] L28H FNR Cluster
Inorganics (Basel), 11, 2023
|
|
1QYU
 
 | |
7N36
 
 | |
7N37
 
 | | Crystal structure of 3-site deamidated variant of human gamma(S)-crystallin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gamma-crystallin S, MAGNESIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Martin, R.W. | | Deposit date: | 2021-05-31 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Deamidation of the human eye lens protein gamma S-crystallin accelerates oxidative aging.
Structure, 30, 2022
|
|
7N38
 
 | | Crystal structure of 5-site deamidated variant of human gamma(S)-crystallin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gamma-crystallin S, MAGNESIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Martin, R.W. | | Deposit date: | 2021-05-31 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Deamidation of the human eye lens protein gamma S-crystallin accelerates oxidative aging.
Structure, 30, 2022
|
|
7N39
 
 | |
7N3A
 
 | |
7N3B
 
 | |
1P11
 
 | |
1P12
 
 | |
6SPA
 
 | | A4V MUTANT OF HUMAN SUPEROXIDE DISMUTASE 1 IN C2 SPACE GROUP | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Shahid, M, Chantadul, V, Amporndanai, K, Wright, G, Antonyuk, S, Hasnain, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy.
Commun Biol, 3, 2020
|
|
1QSZ
 
 | | THE VEGF-BINDING DOMAIN OF FLT-1 (MINIMIZED MEAN) | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 1 | | Authors: | Starovasnik, M.A, Christinger, H.W, Wiesmann, C, Champe, M.A, de Vos, A.M, Skelton, N.J. | | Deposit date: | 1999-06-24 | | Release date: | 1999-11-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the VEGF-binding domain of Flt-1: comparison of its free and bound states.
J.Mol.Biol., 293, 1999
|
|
3P3W
 
 | |
7OEZ
 
 | | Leucine Aminopeptidase A mature enzyme in a complex with leucine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Watson, K.A, Baltulionis, G. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The role of propeptide-mediated autoinhibition and intermolecular chaperone in the maturation of cognate catalytic domain in leucine aminopeptidase.
J.Struct.Biol., 213, 2021
|
|
