1EB4
 
 | |
1EB6
 
 | | Deuterolysin from Aspergillus oryzae | | Descriptor: | 1,2-ETHANEDIOL, NEUTRAL PROTEASE II, ZINC ION | | Authors: | McAuley, K.E, Jia-Xing, Y, Dodson, E.J, Lehmbeck, J, Ostergaard, P.R, Wilson, K.S. | | Deposit date: | 2001-07-19 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Quick Solution: Ab Initio Structure Determination of a 19 kDa Metalloproteinase Using Acorn
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EB7
 
 | |
1EB8
 
 | | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot esculenta | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-ACETONE-CYANOHYDRIN LYASE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Kobler, C, Wajant, H, Effenberger, F. | | Deposit date: | 2001-07-24 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot Esculenta.
Protein Sci., 11, 2002
|
|
1EB9
 
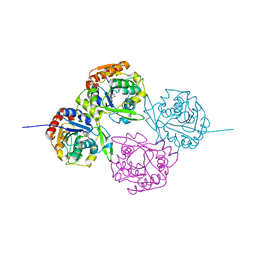 | | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot esculenta | | Descriptor: | HYDROXYNITRILE LYASE, P-HYDROXYBENZALDEHYDE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Kobler, C, Wajant, H, Effenberger, F. | | Deposit date: | 2001-07-24 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot Esculenta.
Protein Sci., 11, 2002
|
|
1EBA
 
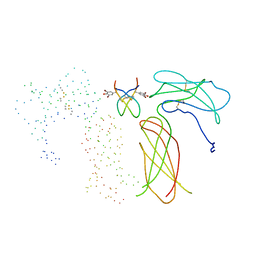 | |
1EBB
 
 | | Bacillus stearothermophilus YhfR | | Descriptor: | GLYCEROL, PHOSPHATASE, SULFATE ION | | Authors: | Rigden, D.J, Jedrzejas, M.J. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Action of a Cofactor-Dependent Phosphoglycerate Mutase Homolog from Bacillus Stearothermophilus with Broad Specificity Phosphatase Activity.
J.Mol.Biol., 315, 2002
|
|
1EBC
 
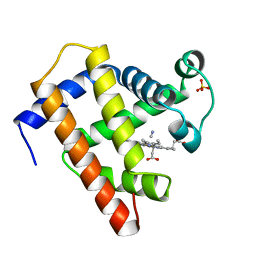 | | SPERM WHALE MET-MYOGLOBIN:CYANIDE COMPLEX | | Descriptor: | CYANIDE ION, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosano, C, Ascenzi, P, Rizzi, M, Losso, R, Bolognesi, M. | | Deposit date: | 1999-03-04 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyanide binding to Lucina pectinata hemoglobin I and to sperm whale myoglobin: an x-ray crystallographic study.
Biophys.J., 77, 1999
|
|
1EBD
 
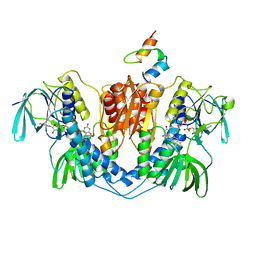 | | DIHYDROLIPOAMIDE DEHYDROGENASE COMPLEXED WITH THE BINDING DOMAIN OF THE DIHYDROLIPOAMIDE ACETYLASE | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE, DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mande, S.S, Sarfaty, S, Allen, M.D, Perham, R.N, Hol, W.G.J. | | Deposit date: | 1996-02-03 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase.
Structure, 4, 1996
|
|
1EBE
 
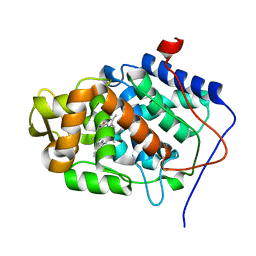 | | Laue diffraction study on the structure of cytochrome c peroxidase compound I | | Descriptor: | CYTOCHROME C PEROXIDASE, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fulop, V, Phizackerley, R.P, Soltis, S.M, Clifton, I.J, Wakatsuki, S, Erman, J.E, Hajdu, J, Edwards, S.L. | | Deposit date: | 2001-07-25 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Laue Diffraction Study on the Structure of Cytochrome C Peroxidase Compound I
Structure, 2, 1994
|
|
1EBF
 
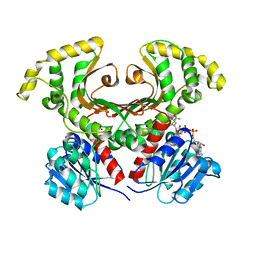 | | HOMOSERINE DEHYDROGENASE FROM S. CEREVISIAE COMPLEX WITH NAD+ | | Descriptor: | HOMOSERINE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | DeLaBarre, B, Thompson, P.R, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
Nat.Struct.Biol., 7, 2000
|
|
1EBG
 
 | |
1EBH
 
 | |
1EBK
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1EBL
 
 | | THE 1.8 A CRYSTAL STRUCTURE AND ACTIVE SITE ARCHITECTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE III (FABH) FROM ESCHERICHIA COLI | | Descriptor: | BETA-KETOACYL-ACP SYNTHASE III, COENZYME A | | Authors: | Davies, C, Heath, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2000-01-24 | | Release date: | 2000-02-11 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from escherichia coli.
Structure Fold.Des., 8, 2000
|
|
1EBM
 
 | | CRYSTAL STRUCTURE OF THE HUMAN 8-OXOGUANINE GLYCOSYLASE (HOGG1) BOUND TO A SUBSTRATE OLIGONUCLEOTIDE | | Descriptor: | 8-OXOGUANINE DNA GLYCOSYLASE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Bruner, S.D, Norman, D.P, Verdine, G.L. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA.
Nature, 403, 2000
|
|
1EBO
 
 | | CRYSTAL STRUCTURE OF THE EBOLA VIRUS MEMBRANE-FUSION SUBUNIT, GP2, FROM THE ENVELOPE GLYCOPROTEIN ECTODOMAIN | | Descriptor: | CHLORIDE ION, EBOLA VIRUS ENVELOPE PROTEIN CHIMERA CONSISTING OF A FRAGMENT OF GCN4 ZIPPER CLONED N-TERMINAL TO A FRAGMENT OF GP2, ZINC ION | | Authors: | Weissenhorn, W, Carfi, A, Lee, K.H, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1998-11-03 | | Release date: | 1999-07-02 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain.
Mol.Cell, 2, 1998
|
|
1EBP
 
 | |
1EBQ
 
 | |
1EBR
 
 | |
1EBS
 
 | |
1EBT
 
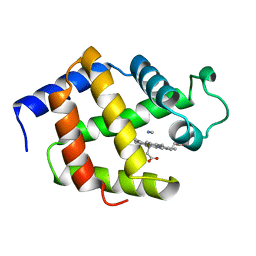 | |
1EBU
 
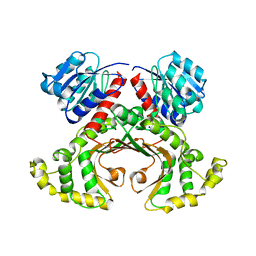 | | HOMOSERINE DEHYDROGENASE COMPLEX WITH NAD ANALOGUE AND L-HOMOSERINE | | Descriptor: | 3-AMINOMETHYL-PYRIDINIUM-ADENINE-DINUCLEOTIDE, HOMOSERINE DEHYDROGENASE, L-HOMOSERINE, ... | | Authors: | DeLaBarre, B, Thompson, P.R, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
Nat.Struct.Biol., 7, 2000
|
|
1EBV
 
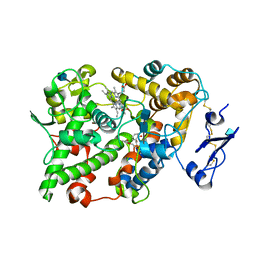 | | OVINE PGHS-1 COMPLEXED WITH SALICYL HYDROXAMIC ACID | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID SALICYLOYL-AMINO-ESTER, ... | | Authors: | Loll, P.J, Sharkey, C.T, O'Connor, S.J, Fitzgerald, D.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-02-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | O-acetylsalicylhydroxamic acid, a novel acetylating inhibitor of prostaglandin H2 synthase: structural and functional characterization of enzyme-inhibitor interactions.
Mol.Pharmacol., 60, 2001
|
|
1EBW
 
 | | HIV-1 protease in complex with the inhibitor BEA322 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DIBENZYL]-GLUCARYL]-DI-[ISOLEUCYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
