5U7G
 
 | | Crystal Structure of the Catalytic Core of CBP | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Park, S, Stanfield, R.L, Martinez-Yamout, M.M, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6PDX
 
 | |
5U2K
 
 | | Crystal structure of Galactoside O-acetyltransferase complex with CoA (H3 space group) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Knapik, A.A, Niedzialkowska, E, Siuda, M.K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-30 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of Galactoside O-acetyltransferase complex
with CoA (H3 space group)
to be published
|
|
4BB7
 
 | | Crystal structure of the yeast Rsc2 BAH domain | | Descriptor: | CHLORIDE ION, CHROMATIN STRUCTURE-REMODELING COMPLEX SUBUNIT RSC2, GLYCEROL, ... | | Authors: | Chambers, A.L, Pearl, L.H, Oliver, A.W, Downs, J.A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bah Domain of Rsc2 is a Histone H3 Binding Domain.
Nucleic Acids Res., 41, 2013
|
|
4BF8
 
 | | Fpr4 PPI domain | | Descriptor: | FPR4 | | Authors: | Monneau, Y, Mackereth, C. | | Deposit date: | 2013-03-15 | | Release date: | 2013-07-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and Activity of the Peptidyl-Prolyl Isomerase Domain from the Histone Chaperone Fpr4 Towards Histone H3 Proline Isomerization
J.Biol.Chem., 288, 2013
|
|
3ZPV
 
 | | Crystal structure of Drosophila Pygo PHD finger in complex with Legless HD1 domain | | Descriptor: | PROTEIN BCL9 HOMOLOG, PROTEIN PYGOPUS, ZINC ION | | Authors: | Miller, T.C.R, Mieszczanek, J, Sanchez-Barrena, M.J, Rutherford, T.J, Fiedler, M, Bienz, M. | | Deposit date: | 2013-03-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Evolutionary Adaptation of the Fly Pygo Phd Finger Towards Recognizing Histone H3 Tail Methylated at Arginine 2
Structure, 21, 2013
|
|
5E6H
 
 | | A Linked Jumonji Domain of the KDM5A Lysine Demethylase | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2015-10-09 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Characterization of a Linked Jumonji Domain of the KDM5/JARID1 Family of Histone H3 Lysine 4 Demethylases.
J.Biol.Chem., 291, 2016
|
|
3LX7
 
 | | Crystal structure of a Novel Tudor domain-containing protein SGF29 | | Descriptor: | SAGA-associated factor 29 homolog, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Bian, C.B, Xu, C, Tempel, W, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3LJW
 
 | | Crystal Structure of the Second Bromodomain of Human Polybromo | | Descriptor: | ACETATE ION, Protein polybromo-1, SODIUM ION | | Authors: | Charlop-Powers, Z, Zhou, M.M, Zeng, L, Zhang, Q. | | Deposit date: | 2010-01-26 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2.
Cell Res., 20, 2010
|
|
7ZMX
 
 | |
3MP6
 
 | | Complex Structure of Sgf29 and dimethylated H3K4 | | Descriptor: | H3K4me2 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, J, Wu, M, Ruan, J, Zang, J. | | Deposit date: | 2010-04-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
3MEW
 
 | | Crystal structure of Novel Tudor domain-containing protein SGF29 | | Descriptor: | SAGA-associated factor 29 homolog | | Authors: | Xu, C, Bian, C.B, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3MP8
 
 | | Crystal structure of Sgf29 tudor domain | | Descriptor: | 4-(HYDROXYMETHYL)BENZAMIDINE, ACETIC ACID, GLYCEROL, ... | | Authors: | Li, J, Wu, M, Ruan, J, Zang, J. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
3MUG
 
 | | Crystal structure of human Fab PG16, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody PG16 Heavy Chain, ... | | Authors: | Pejchal, R, Walker, L.M, Burton, D.R, Wilson, I.A. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MUH
 
 | | Crystal structure of PG9 light chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody PG9 light chain | | Authors: | Pejchal, R, Walker, L.M, Burton, D.R, Wilson, I.A. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7E7C
 
 | |
2EH8
 
 | | Crystal structure of the complex of humanized KR127 fab and PRES1 peptide epitope | | Descriptor: | HUMANIZED KR127 FAB, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Chi, S.-W, Kim, S.-J, Maeng, C.-Y, Hong, H.J, Ryu, S.-E. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2RR6
 
 | | Solution structure of the leucine rich repeat of human acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Umehara, T, Tsuda, K, Koshiba, S, Harada, T, Watanabe, S, Tanaka, A, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
2ELL
 
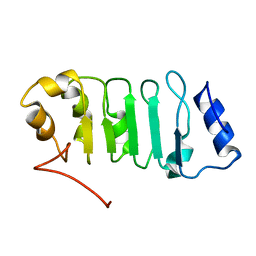 | | Solution structure of the Leucine Rich Repeat of human Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
2DLF
 
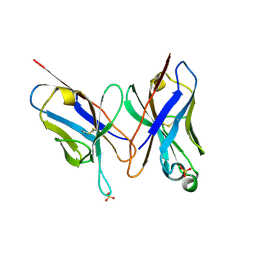 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 6.75 | | Descriptor: | PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S) (HEAVY CHAIN)), PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S)-KAPPA (LIGHT CHAIN)), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2EH7
 
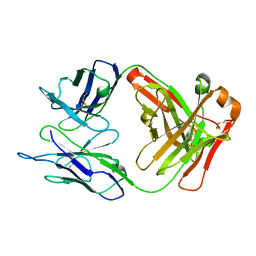 | | Crystal structure of humanized KR127 FAB | | Descriptor: | HUMANIZED KR127 FAB, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Chi, S.-W, Kim, S.-J, Maeng, C.-Y, Hong, H.J, Ryu, S.-E. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | Descriptor: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | Authors: | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
5TRL
 
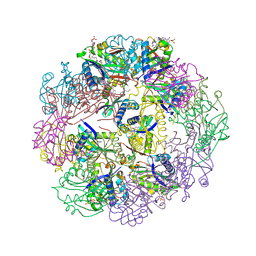 | |
5TRM
 
 | |
7F4A
 
 | |
