7YCE
 
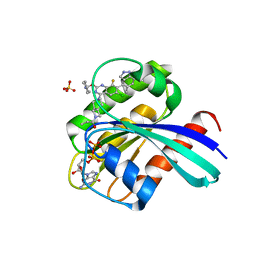 | | KRas G12C in complex with Compound 7b | | Descriptor: | 1-[7-[6-chloranyl-2-(1-ethylpiperidin-4-yl)oxy-8-fluoranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Amano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and biological evaluation of 1-{2,7-diazaspiro[3.5]nonan-2-yl}prop-2-en-1-one derivatives as covalent inhibitors of KRAS G12C with favorable metabolic stability and anti-tumor activity.
Bioorg.Med.Chem., 71, 2022
|
|
7YCC
 
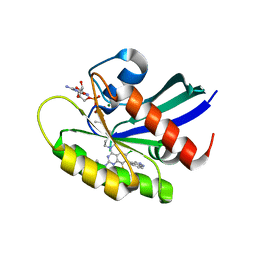 | | KRas G12C in complex with Compound 5c | | Descriptor: | 1-[7-[6-chloranyl-8-fluoranyl-7-(5-methyl-1~{H}-indazol-4-yl)-2-[(1-methylpiperidin-4-yl)amino]quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Amano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and biological evaluation of 1-{2,7-diazaspiro[3.5]nonan-2-yl}prop-2-en-1-one derivatives as covalent inhibitors of KRAS G12C with favorable metabolic stability and anti-tumor activity.
Bioorg.Med.Chem., 71, 2022
|
|
6T0F
 
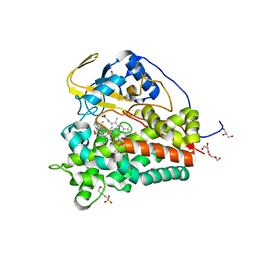 | | Crystal structure of CYP124 in complex with cholest-4-en-3-one | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
5JVD
 
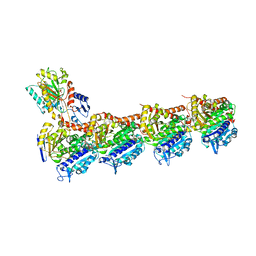 | | Tubulin-TUB092 complex | | Descriptor: | (2E)-3-(3-hydroxy-4-methoxyphenyl)-1-(7-methoxy-2H-1,3-benzodioxol-5-yl)-2-methylprop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Canela, M.-D, Noppen, S, Bueno, O, Prota, A.E, Bargsten, K, Saez-Calvo, G, Jimeno, M.-L, Benkheil, M, Ribatti, D, Velazquez, S, Camarasa, M.-J, Diaz, J.F, Steinmetz, M.O, Priego, E.-M, Perez-Perez, M.-J, Liekens, S. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Antivascular and antitumor properties of the tubulin-binding chalcone TUB091.
Oncotarget, 8, 2017
|
|
5N4Z
 
 | |
4MWE
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[5-chloro-3-(prop-2-en-1-yl)-2-(prop-2-en-1-yloxy)benzyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1475) | | Descriptor: | 1-(3-{[5-chloro-3-(prop-2-en-1-yl)-2-(prop-2-en-1-yloxy)benzyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4KVY
 
 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (1S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1S,5S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Al-lami, N, Janvier, M, D'Antonio, E.L, Faraldos, J.A, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4KWD
 
 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (1R,8R,9aS)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1R,5R,8R,9aS)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Al-lami, N, Janvier, M, D'Antonio, E.L, Faraldos, J.A, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4KVW
 
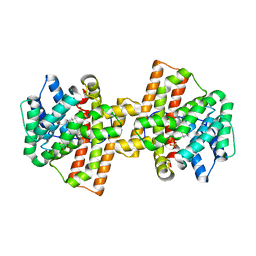 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (3R,6R,9aR)-6,9a-dimethyl-3-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (3R,5S,6R,9aR)-6,9a-dimethyl-3-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Al-lami, N, Janvier, M, D'Antonio, E.L, Faraldos, J.A, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
5TML
 
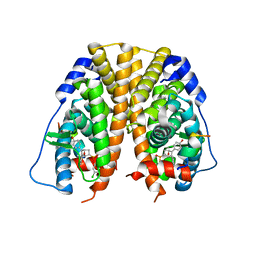 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, (E)-6-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | Descriptor: | 6-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
1QJT
 
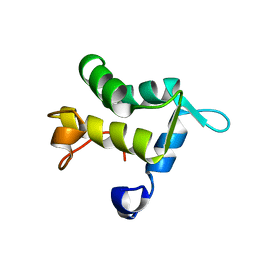 | | SOLUTION STRUCTURE OF THE APO EH1 DOMAIN OF MOUSE EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE 15, EPS15 | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE SUBSTRATE 15, EPS15 | | Authors: | Whitehead, B, Tessari, M, Carotenuto, A, van Bergen en Henegouwen, P.M, Vuister, G.W. | | Deposit date: | 1999-07-02 | | Release date: | 2000-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eh1 Domain of Eps15 is Structurally Classified as a Member of the S100 Subclass of EF-Hand Containing Proteins
Biochemistry, 38, 1999
|
|
7B0Z
 
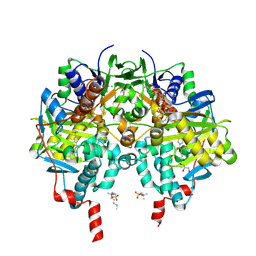 | |
7B0V
 
 | |
6T9F
 
 | | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.24847341 Å) | | Cite: | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM
To Be Published
|
|
3ZGL
 
 | | Crystal structures of Escherichia coli IspH in complex with AMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-4-amino-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
3ZGN
 
 | | Crystal structures of Escherichia coli IspH in complex with TMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-3-methyl-4-sulfanylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
7SMZ
 
 | | X-ray crystal structure of CYP142A3 from Mycobacterium Marinum in complex with 4-cholesten-3-one | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, ACETATE ION, Cytochrome P450 142A3, ... | | Authors: | Ghith, A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2021-10-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Structures of the Steroid Binding CYP142 Cytochrome P450 Enzymes from Mycobacterium ulcerans and Mycobacterium marinum.
Acs Infect Dis., 8, 2022
|
|
5TM6
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)hexanoic acid | | Descriptor: | 6-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}hexanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TLF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Constrained WAY Derivative, 4-(2-(3-methylbut-2-en-1-yl)-7-(trifluoromethyl)-2H-indazol-3-yl)benzene-1,3-diol | | Descriptor: | 4-[2-(3-methylbut-2-en-1-yl)-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, NUCLEAR RECEPTOR COACTIVATOR 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-11 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, 5-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)pentanoic acid | | Descriptor: | 5-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}pentanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC analog, (E)-6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | Descriptor: | 6-{4-[(1S,4S,6S)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM7
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | Descriptor: | 7-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM8
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | Descriptor: | 7-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, (E)-3-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)acrylic acid | | Descriptor: | 3-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}prop-2-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
7LOB
 
 | | T4 lysozyme mutant L99A in complex with 1-fluoro-2-[(prop-2-en-1-yl)oxy]benzene | | Descriptor: | 1-fluoro-2-[(prop-2-en-1-yl)oxy]benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
