8ADG
 
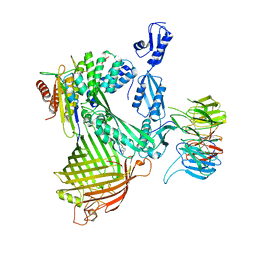 | | Cryo-EM structure of Darobactin 22 bound BAM complex | | Descriptor: | Darobactin 22, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7PT2
 
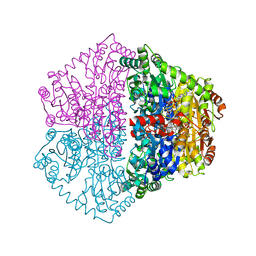 | |
7PT3
 
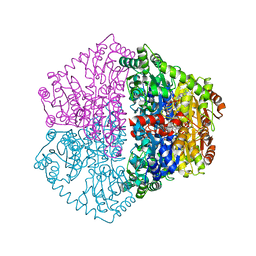 | |
7PT1
 
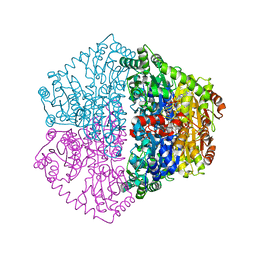 | |
7PT4
 
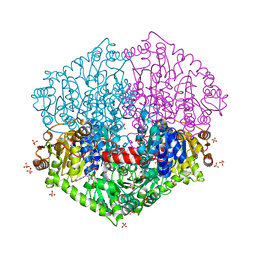 | | Actinobacterial 2-hydroxyacyl-CoA lyase (AcHACL) structure in complex with a covalently bound reaction intermediate as well as products formyl-CoA and acetone | | Descriptor: | 2-hydroxyacyl-CoA lyase, 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1R,11R,15S,17R)-19-[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]-1,11,15,17-TETRAHYDROXY-12,12-DIMETHYL-15,17-DIOXIDO-6,10-DIOXO-14,16,18-TRIOXA-2-THIA-5,9-DIAZA-15,17-DIPHOSPHANONADEC-1-YL}-5-(2-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, ACETONE, ... | | Authors: | Zahn, M, Rohwerder, T. | | Deposit date: | 2021-09-25 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanistic details of the actinobacterial lyase-catalyzed degradation reaction of 2-hydroxyisobutyryl-CoA.
J.Biol.Chem., 298, 2022
|
|
8BVQ
 
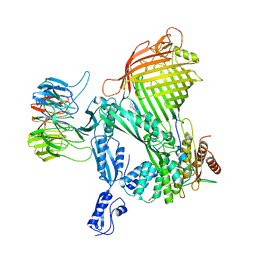 | | E. coli BAM complex (BamABCDE) bound to darobactin B | | Descriptor: | Darobactin-B, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Horne, J.E, Fenn, K.L, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Darobactin B Stabilises a Lateral-Closed Conformation of the BAM Complex in E. coli Cells.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1LRY
 
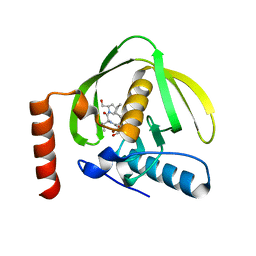 | | Crystal Structure of P. aeruginosa Peptide Deformylase Complexed with Antibiotic Actinonin | | Descriptor: | ACTINONIN, PEPTIDE deformylase, ZINC ION | | Authors: | Guilloteau, J.-P, Mathieu, M, Giglione, C, Blanc, V, Dupuy, A, Chevrier, M, Gil, P, Famechon, A, Meinnel, T, Mikol, V. | | Deposit date: | 2002-05-16 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
1LQY
 
 | |
1LRU
 
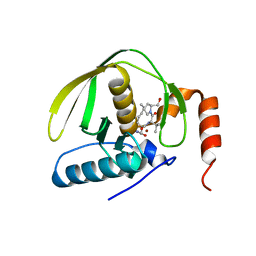 | | Crystal Structure of E.coli Peptide Deformylase Complexed with Antibiotic Actinonin | | Descriptor: | ACTINONIN, PEPTIDE DEFORMYLASE, SULFATE ION, ... | | Authors: | Guilloteau, J.-P, Mathieu, M, Giglione, C, Blanc, V, Dupuy, A, Chevrier, M, Gil, P, Famechon, A, Meinnel, T, Mikol, V. | | Deposit date: | 2002-05-16 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
2KMN
 
 | |
3A07
 
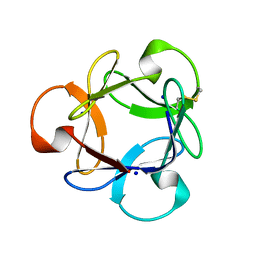 | | Crystal Structure of Actinohivin; Potent anti-HIV Protein | | Descriptor: | Actinohivin, SODIUM ION | | Authors: | Tsunoda, M, Suzuki, K, Sagara, T, Takenaka, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-08-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Mechanism by which the lectin actinohivin blocks HIV infection of target cells
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1ACX
 
 | |
1J8I
 
 | | Solution Structure of Human Lymphotactin | | Descriptor: | Lymphotactin | | Authors: | Kuloglu, E.S, McCaslin, D.R, Markley, J.L, Pauza, C.D, Volkman, B.F. | | Deposit date: | 2001-05-21 | | Release date: | 2001-10-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
1J9O
 
 | | SOLUTION STRUCTURE OF HUMAN LYMPHOTACTIN | | Descriptor: | LYMPHOTACTIN | | Authors: | Kuloglu, E.S, McCaslin, D.R, Kitabwalla, M, Pauza, C.D, Markley, J.L, Volkman, B.F. | | Deposit date: | 2001-05-28 | | Release date: | 2001-10-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
1TJT
 
 | |
8P94
 
 | |
3J8K
 
 | | Tilted state of actin, T2 | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Galkin, V.E, Orlova, A, Vos, M.R, Schroder, G.F, Egelman, E.H. | | Deposit date: | 2014-11-07 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Near-atomic resolution for one state of f-actin.
Structure, 23, 2015
|
|
3J8J
 
 | | Tilted state of actin, T1 | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Galkin, V.E, Orlova, A, Vos, M.R, Schroder, G.F, Egelman, E.H. | | Deposit date: | 2014-11-07 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Near-atomic resolution for one state of f-actin.
Structure, 23, 2015
|
|
7U8K
 
 | | Magic Angle Spinning NMR Structure of Human Cofilin-2 Assembled on Actin Filaments | | Descriptor: | Actin, alpha skeletal muscle, Cofilin-2 | | Authors: | Kraus, J, Russell, R, Kudryashova, E, Xu, C, Katyal, N, Kudryashov, D, Perilla, J.R, Polenova, T. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | SOLID-STATE NMR | | Cite: | Magic angle spinning NMR structure of human cofilin-2 assembled on actin filaments reveals isoform-specific conformation and binding mode.
Nat Commun, 13, 2022
|
|
1PEV
 
 | | Crystal Structure of the Actin Interacting Protein from Caenorhabditis Elegans | | Descriptor: | Actin interacting protein 1 | | Authors: | Vorobiev, S, Mohri, K, Fedorov, A.A, Ono, S, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-05-22 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments.
J.Biol.Chem., 279, 2004
|
|
2A41
 
 | | Ternary complex of the WH2 Domain of WIP with Actin-DNAse I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chereau, D, Kerff, F, Dominguez, R. | | Deposit date: | 2005-06-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1NR0
 
 | | Two Seven-Bladed Beta-Propeller Domains Revealed By The Structure Of A C. elegans Homologue Of Yeast Actin Interacting Protein 1 (AIP1). | | Descriptor: | Actin interacting protein 1, MANGANESE (II) ION | | Authors: | Vorobiev, S.M, Mohri, K, Ono, S, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-23 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of Functional Residues on Caenorhabditis elegans Actin-interacting Protein 1 (UNC-78) for Disassembly of Actin Depolymerizing Factor/Cofilin-bound Actin Filaments
J.Biol.Chem., 279, 2004
|
|
5LY3
 
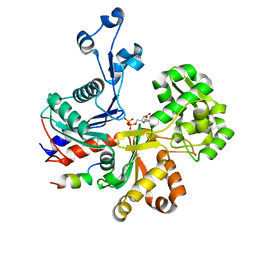 | |
1PGU
 
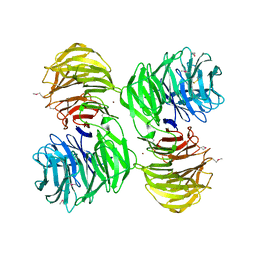 | | YEAST ACTIN INTERACTING PROTEIN 1 (AIP1), Se-Met PROTEIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | Actin interacting protein 1, ZINC ION | | Authors: | Voegtli, W.C, Madrona, A.Y, Wilson, D.K. | | Deposit date: | 2003-05-28 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization.
J.Biol.Chem., 278, 2003
|
|
1PI6
 
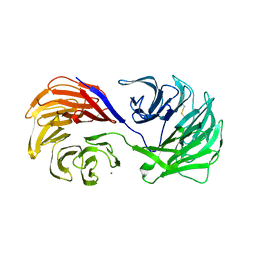 | | YEAST ACTIN INTERACTING PROTEIN 1 (Aip1), ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | Actin interacting protein 1, ZINC ION | | Authors: | Voegtli, W.C, Madrona, A.Y, Wilson, D.K. | | Deposit date: | 2003-05-29 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization.
J.Biol.Chem., 278, 2003
|
|
