7LKJ
 
 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) | | Descriptor: | 1,2-ETHANEDIOL, Aminofutalosine deaminase, FE (III) ION | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
7LKK
 
 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) in complex with Methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, 1,2-ETHANEDIOL, Aminofutalosine deaminase, ... | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
4V1X
 
 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WHB
 
 | | Crystal structure of phenylurea hydrolase B | | Descriptor: | Phenylurea hydrolase B, ZINC ION | | Authors: | Sugrue, E, Carr, P.D, Khurana, J.L, Jackson, C.J. | | Deposit date: | 2014-09-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
4WGX
 
 | | Crystal Structure of Molinate Hydrolase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COBALT (II) ION, Molinate hydrolase | | Authors: | Sugrue, E, Carr, P.D, Fraser, N.J, Hopkins, D.H, Jackson, C.J. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
4YIW
 
 | | DIHYDROOROTASE FROM BACILLUS ANTHRACIS WITH SUBSTRATE BOUND | | Descriptor: | Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ZINC ION | | Authors: | Lei, H, Santarsiero, B.D, Rice, A.J, Lee, H, Johnson, M.E. | | Deposit date: | 2015-03-02 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Ca-asp bound X-ray structure and inhibition of Bacillus anthracis dihydroorotase (DHOase).
Bioorg.Med.Chem., 24, 2016
|
|
4TQT
 
 | |
6OH9
 
 | | Yeast Guanine Deaminase | | Descriptor: | NONAETHYLENE GLYCOL, SULFATE ION, Yeast Guanine Deaminase, ... | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
6OHC
 
 | | E. coli Guanine Deaminase | | Descriptor: | GLYCEROL, Guanine deaminase, ZINC ION | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
6OHB
 
 | | E. coli Guanine Deaminase | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
6OHA
 
 | | Yeast Guanine Deaminase | | Descriptor: | PENTAETHYLENE GLYCOL, Probable guanine deaminase, SULFATE ION, ... | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
5LP3
 
 | |
3V7P
 
 | | Crystal structure of amidohydrolase nis_0429 (target efi-500396) from Nitratiruptor sp. sb155-2 | | Descriptor: | Amidohydrolase family protein, BENZOIC ACID, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-21 | | Release date: | 2012-01-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Amidohydrolase Nis_0429 (Target Efi-500319) from Nitratiruptor Sp. Sb155-2
To be Published
|
|
5MKV
 
 | | Crystal Structure of Human Dihydropyrimidinease-like 2 (DPYSL2A)/Collapsin Response Mediator Protein (CRMP2) residues 13-516 | | Descriptor: | 1,2-ETHANEDIOL, Dihydropyrimidinase-related protein 2 | | Authors: | Sethi, R, Zheng, Y, Krojer, T, Velupillai, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ahmed, A.A, von Delft, F. | | Deposit date: | 2016-12-05 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tuning microtubule dynamics to enhance cancer therapy by modulating FER-mediated CRMP2 phosphorylation.
Nat Commun, 9, 2018
|
|
5NKS
 
 | | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4) | | Descriptor: | Dihydropyrimidinase-related protein 4 | | Authors: | Mathea, S, Elkins, J.M, Strain-Damerell, C, Salah, E, Borkowska, O, Chalk, R, Burgess-Brown, N, Pinkas, D.M, von Delft, F, Krojer, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2017-03-31 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4)
To Be Published
|
|
5MLE
 
 | | Crystal Structure of Human Dihydropyrimidinease-like 2 (DPYSL2A)/Collapsin Response Mediator Protein (CRMP2 13-516) Mutant Y479E/Y499E | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dihydropyrimidinase-related protein 2, ... | | Authors: | Sethi, R, Zheng, Y, Talon, R, Velupillai, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ahmed, A.A, von Delft, F. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Tuning microtubule dynamics to enhance cancer therapy by modulating FER-mediated CRMP2 phosphorylation.
Nat Commun, 9, 2018
|
|
5LXX
 
 | | High-resolution structure of human collapsin response mediator protein 2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydropyrimidinase-related protein 2, SULFATE ION | | Authors: | Myllykoski, M, Hensley, K, Kursula, P. | | Deposit date: | 2016-09-23 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Collapsin response mediator protein 2: high-resolution crystal structure sheds light on small-molecule binding, post-translational modifications, and conformational flexibility.
Amino Acids, 49, 2017
|
|
4AQL
 
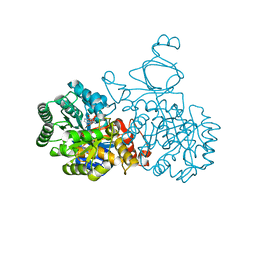 | | HUMAN GUANINE DEAMINASE IN COMPLEX WITH VALACYCLOVIR | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl L-valinate, GUANINE DEAMINASE, ZINC ION | | Authors: | Welin, M, Egeblad, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, Weigelt, J, Nordlund, P. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Pan-Pathway Based Interaction Profiling of Fda-Approved Nucleoside and Nucleobase Analogs with Enzymes of the Human Nucleotide Metabolism.
Plos One, 7, 2012
|
|
4B91
 
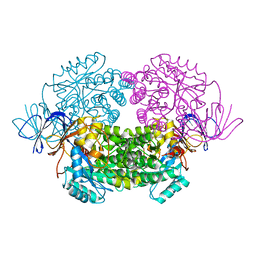 | | Crystal structure of truncated human CRMP-5 | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 5 | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights Into the Oligomerization of Crmps: Crystal Structure of Human Collapsin Response Mediator Protein 5.
J.Neurochem., 125, 2013
|
|
4B90
 
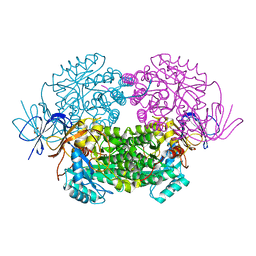 | | Crystal structure of WT human CRMP-5 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-RELATED PROTEIN 5 | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights Into the Oligomerization of Crmps: Crystal Structure of Human Collapsin Response Mediator Protein 5.
J.Neurochem., 125, 2013
|
|
3E74
 
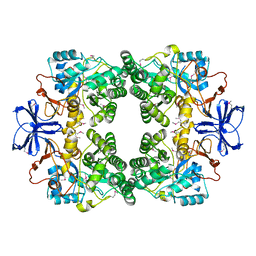 | |
3EGJ
 
 | | N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae. | | Descriptor: | N-acetylglucosamine-6-phosphate deacetylase, NICKEL (II) ION, SULFATE ION | | Authors: | Osipiuk, J, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae.
To be Published
|
|
3SFW
 
 | |
2I9U
 
 | |
2VUN
 
 | | The Crystal Structure of Enamidase at 1.9 A Resolution - A new Member of the Amidohydrolase Superfamily | | Descriptor: | CHLORIDE ION, ENAMIDASE, FE (III) ION, ... | | Authors: | Kress, D, Alhapel, A, Pierik, A.J, Essen, L.-O. | | Deposit date: | 2008-05-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure of Enamidase: A Bifunctional Enzyme of the Nicotinate Catabolism.
J.Mol.Biol., 384, 2008
|
|
