2K3K
 
 | |
2JQU
 
 | |
2K2D
 
 | | Solution NMR structure of C-terminal domain of human pirh2. Northeast Structural Genomics Consortium (NESG) target HT2C | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, ZINC ION | | Authors: | Lemak, A, Sheng, Y, Karra, M, Srisailam, S, Laister, R.C, Duan, S, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of Pirh2-mediated p53 ubiquitylation.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2K2C
 
 | | Solution NMR structure of N-terminal domain of human pirh2. Northeast Structural Genomics Consortium (NESG) target HT2A | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, ZINC ION | | Authors: | Wu, B, Lemak, A, Sheng, Y, Karra, M, Srisailam, S, Sunnerhagen, M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of Pirh2-mediated p53 ubiquitylation.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2JQS
 
 | |
2BDO
 
 | | SOLUTION STRUCTURE OF HOLO-BIOTINYL DOMAIN FROM ACETYL COENZYME A CARBOXYLASE OF ESCHERICHIA COLI DETERMINED BY TRIPLE-RESONANCE NMR SPECTROSCOPY | | Descriptor: | BIOTIN, PROTEIN (ACETYL-COA CARBOXYLASE) | | Authors: | Roberts, E.L, Shu, N, Howard, M.J, Broadhurst, R.W, Chapman-Smith, A, Wallace, J.C, Morris, T, Cronan, J.E, Perham, R.N. | | Deposit date: | 1999-03-03 | | Release date: | 1999-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
2EZH
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZI
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, 30 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZK
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, REGULARIZED MEAN STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-10-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2EZL
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, 29 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-10-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2JR5
 
 | | Solution structure of UPF0350 protein VC_2471. Northeast Structural Genomics Target VcR36 | | Descriptor: | UPF0350 protein VC_2471 | | Authors: | Wu, Y, Parish, D, Singarapu, K.K, Sukumaran, D, Eletski, A, Shastry, R, Nwosu, C, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-20 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of UPF0350 protein VC_2471.
To be Published
|
|
2JY0
 
 | |
1X9A
 
 | | Solution NMR Structure of Protein Tm0979 from Thermotoga maritima. Ontario Center for Structural Proteomics Target TM0979_1_87; Northeast Structural Genomics Consortium Target VT98. | | Descriptor: | hypothetical protein TM0979 | | Authors: | Gaspar, J.A, Liu, C, Vassall, K.A, Stathopulos, P.B, Meglei, G, Stephen, R, Pineda-Lucena, A, Wu, B, Yee, A, Arrowsmith, C.H, Meiering, E.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel member of the YchN-like fold: solution structure of the hypothetical protein Tm0979 from Thermotoga maritima.
Protein Sci., 14, 2005
|
|
2C0W
 
 | | Molecular Structure of fd Filamentous Bacteriophage Refined with Respect to X-ray Fibre Diffraction | | Descriptor: | COAT PROTEIN B | | Authors: | Marvin, D.A, Welsh, L.C, Symmons, M.F, Scott, W.R.P, Straus, S.K. | | Deposit date: | 2005-09-08 | | Release date: | 2005-12-14 | | Last modified: | 2024-02-14 | | Method: | FIBER DIFFRACTION (3.2 Å) | | Cite: | Molecular Structure of Fd (F1, M13) Filamentous Bacteriophage Refined with Respect to X-Ray Fibre Diffraction and Solid-State NMR Data Supports Specific Models of Phage Assembly at the Bacterial Membrane.
J.Mol.Biol., 355, 2006
|
|
2BPN
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERRICYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Aguiar, A.P, Brennan, L, Xavier, A.V, Turner, D.L. | | Deposit date: | 2005-04-21 | | Release date: | 2006-03-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Tetrahaem Ferricytochrome C(3) from Desulfovibrio Vulgaris (Hildenborough) and its K45Q Mutant: The Molecular Basis of Cooperativity.
Biochim.Biophys.Acta, 1757, 2006
|
|
2J6D
 
 | | CONKUNITZIN-S2 - CONE SNAIL NEUROTOXIN - DENOVO STRUCTURE | | Descriptor: | CONKUNITZIN-S2 | | Authors: | Korukottu, J, Bayrhuber, M, Montaville, P, Vijayan, V, Jung, Y.-S, Becker, S, Zweckstetter, M. | | Deposit date: | 2006-09-27 | | Release date: | 2007-01-16 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | Fast High-Resolution Protein Structure Determination by Using Unassigned NMR Data.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2BB8
 
 | |
2KOJ
 
 | |
2AXX
 
 | | THE SOLUTION STRUCTURE OF OXIDIZED RAT MICROSOMAL CYTOCHROME B5, NMR, 21 STRUCTURES | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized rat microsomal cytochrome b5.
Biochemistry, 37, 1998
|
|
2KNB
 
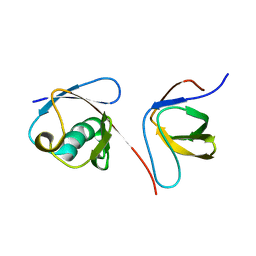 | | Solution NMR structure of the parkin Ubl domain in complex with the endophilin-A1 SH3 domain | | Descriptor: | E3 ubiquitin-protein ligase parkin, Endophilin-A1 | | Authors: | Trempe, J, Guennadi, K, Edna, C.M, Kalle, G. | | Deposit date: | 2009-08-20 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination.
Mol.Cell, 36, 2009
|
|
1YSG
 
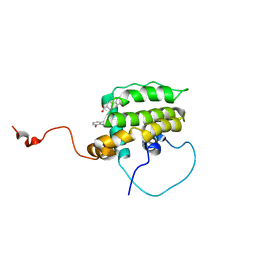 | | Solution Structure of the Anti-apoptotic Protein Bcl-xL in Complex with "SAR by NMR" Ligands | | Descriptor: | 4'-FLUORO-1,1'-BIPHENYL-4-CARBOXYLIC ACID, 5,6,7,8-TETRAHYDRONAPHTHALEN-1-OL, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1ZZJ
 
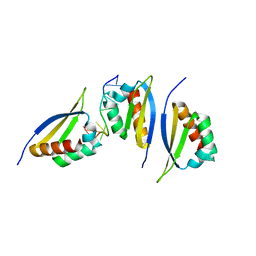 | | Structure of the third KH domain of hnRNP K in complex with 15-mer ssDNA | | Descriptor: | 5'-D(*TP*TP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*TP*TP*T)-3', Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
1ZZK
 
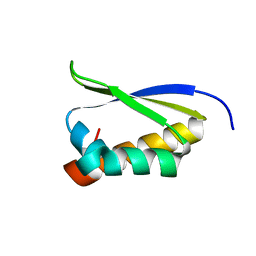 | | Crystal Structure of the third KH domain of hnRNP K at 0.95A resolution | | Descriptor: | Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
2EZP
 
 | |
2KZ8
 
 | | Solution NMR structure of MqsA, a protein from E. coli, containing a Zinc finger, N-terminal and a Helix Turn-Helix C-terminal domain | | Descriptor: | Uncharacterized HTH-type transcriptional regulator ygiT | | Authors: | Papadopoulos, E, Vlamis-Gardikas, A, Graslund, A, Billeter, M, Holmgren, A, Collet, J. | | Deposit date: | 2010-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biophysical properties of MqsA, a Zn-containing antitoxin from Escherichia coli
Biochim.Biophys.Acta, 2012
|
|
