4GMJ
 
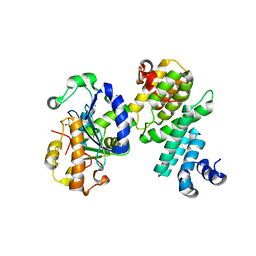 | | Structure of human NOT1 MIF4G domain co-crystallized with CAF1 | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 7, CHLORIDE ION, ... | | Authors: | Petit, P, Weichenrieder, O, Wohlbold, L, Izaurralde, E. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex
Nucleic Acids Res., 40, 2012
|
|
6KWB
 
 | |
6L29
 
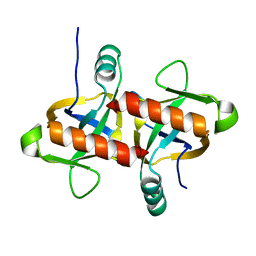 | | The structure of the MazF-mt1 mutant | | Descriptor: | mRNA interferase | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-10-02 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3000052 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6L5B
 
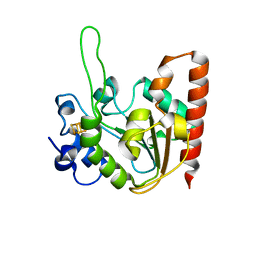 | | The structure of the UdgX mutant H109E at a post-excision state | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Xie, W, Tu, J, Zeng, H. | | Deposit date: | 2019-10-22 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.00004983 Å) | | Cite: | Structural insights into an MsmUdgX mutant capable of both crosslinking and uracil excision capability.
DNA Repair (Amst), 97, 2021
|
|
2CH5
 
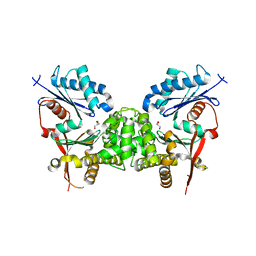 | | Crystal structure of human N-acetylglucosamine kinase in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Weihofen, W.A, Berger, M, Chen, H, Saenger, W, Hinderlich, S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-09-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with Adp/Glucose: Insights Into Substrate Specificity and Regulation.
J.Mol.Biol., 364, 2006
|
|
7PQH
 
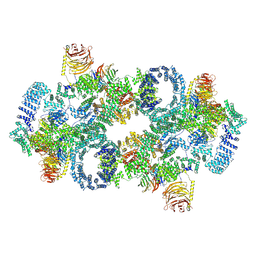 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4GVM
 
 | |
3F62
 
 | |
3KM6
 
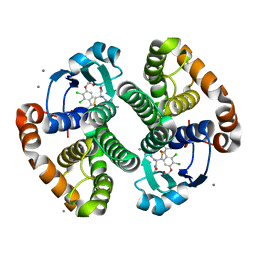 | |
2CH6
 
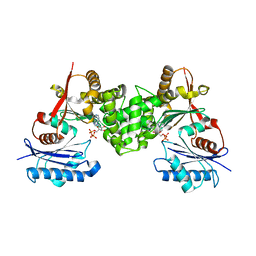 | | Crystal structure of human N-acetylglucosamine kinase in complex with ADP and glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N-ACETYL-D-GLUCOSAMINE KINASE, alpha-D-glucopyranose | | Authors: | Weihofen, W.A, Berger, M, Chen, H, Saenger, W, Hinderlich, S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with Adp/Glucose: Insights Into Substrate Specificity and Regulation.
J.Mol.Biol., 364, 2006
|
|
3EE9
 
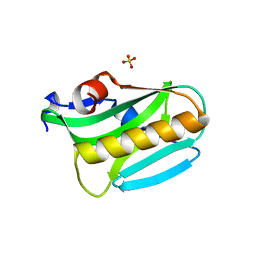 | | Structure of NS1 effector domain | | Descriptor: | Non-structural protein 1, SULFATE ION | | Authors: | Xia, S, Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2008-09-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of NS1A effector domain from the influenza A/Udorn/72 virus.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5CHT
 
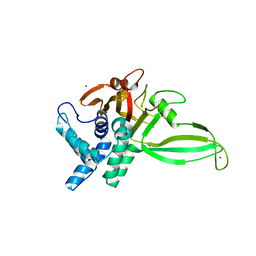 | | Crystal structure of USP18 | | Descriptor: | Ubl carboxyl-terminal hydrolase 18, ZINC ION | | Authors: | Fritz, G, Basters, A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of the specificity of USP18 toward ISG15.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3EE8
 
 | |
5YZW
 
 | | Crystal structure of p204 HINb domain | | Descriptor: | Ifi204 | | Authors: | Jin, T. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
2DFS
 
 | | 3-D structure of Myosin-V inhibited state | | Descriptor: | Calmodulin, Myosin-5A | | Authors: | Liu, J, Taylor, D.W, Krementsova, E.B, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (24 Å) | | Cite: | Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography
Nature, 442, 2006
|
|
3PKV
 
 | |
5KDU
 
 | | ZmpB metallopeptidase in complex with a2,6-Sialyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, SERINE, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KAH
 
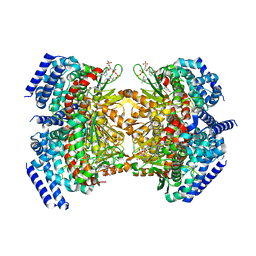 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21, V425T mutant | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.779 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5KAG
 
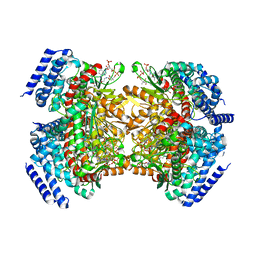 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21 | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5KDS
 
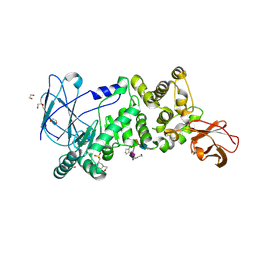 | | ZmpB metallopeptidase in complex with an O-glycopeptide (a2,6-sialylated core-3 pentapeptide). | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6O84
 
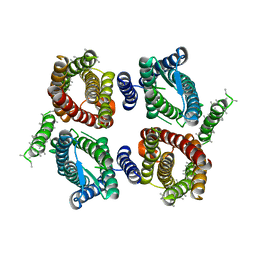 | | Cryo-EM structure of OTOP3 from xenopus tropicalis | | Descriptor: | LOC100127796 protein,LOC100127796 protein,OTOP3,LOC100127796 protein,LOC100127796 protein | | Authors: | Chen, Q.F, Bai, X.C, Jiang, Y.X. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural and functional characterization of an otopetrin family proton channel.
Elife, 8, 2019
|
|
5KOL
 
 | | Crystal structure of the competence-damaged protein (CinA) superfamily protein ECK1530/EC0983 from Escherichia coli | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Wawrak, Z, Evdokimova, E, Di Leo, R, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-30 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | To be published
To Be Published
|
|
2BE6
 
 | | 2.0 A crystal structure of the CaV1.2 IQ domain-Ca/CaM complex | | Descriptor: | CALCIUM ION, Calmodulin 2, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, Chatelain, F.C, Minor Jr, D.L. | | Deposit date: | 2005-10-23 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into voltage-gated calcium channel regulation from the structure of the Ca(V)1.2 IQ domain-Ca(2+)/calmodulin complex
Nat.Struct.Mol.Biol., 12, 2005
|
|
7KTS
 
 | | Negative stain EM structure of the human SAGA coactivator complex (TRRAP, core, splicing module) | | Descriptor: | Ataxin-7, Isoform 3 of Transcription factor SPT20 homolog, STAGA complex 65 subunit gamma, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (19.09 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2I5B
 
 | | The crystal structure of an ADP complex of Bacillus subtilis pyridoxal kinase provides evidence for the parralel emergence of enzyme activity during evolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphomethylpyrimidine kinase | | Authors: | Newman, J.A, Das, S.K, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of an ADP Complex of Bacillus subtilis Pyridoxal Kinase Provides Evidence for the Parallel Emergence of Enzyme Activity During Evolution.
J.Mol.Biol., 363, 2006
|
|
