2PT9
 
 | | The structure of Plasmodium falciparum spermidine synthase in complex with decarboxylated S-adenosylmethionine and the inhibitor cis-4-methylcyclohexylamine (4MCHA) | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, GLYCEROL, ... | | Authors: | Dufe, V.T, Qiu, W, Muller, I.B, Hui, R, Walter, R.D, Al-Karadaghi, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Plasmodium falciparum spermidine synthase in complex with the substrate decarboxylated S-adenosylmethionine and the potent inhibitors 4MCHA and AdoDATO.
J.Mol.Biol., 373, 2007
|
|
2PTA
 
 | | PANDINUS TOXIN K-A (PITX-KA) FROM PANDINUS IMPERATOR, NMR, 20 STRUCTURES | | Descriptor: | PANDINUS TOXIN K-ALPHA | | Authors: | Tenenholz, T.C, Rogowski, R.S, Collins, J.H, Blaustein, M.P, Weber, D.J. | | Deposit date: | 1996-11-26 | | Release date: | 1997-12-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure for Pandinus toxin K-alpha (PiTX-K alpha), a selective blocker of A-type potassium channels.
Biochemistry, 36, 1997
|
|
2PTC
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, TRYPSIN INHIBITOR | | Authors: | Huber, R, Deisenhofer, J. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
2PTD
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT D198E | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-16 | | Release date: | 1998-01-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
2PTF
 
 | | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Uncharacterized protein MTH_863 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN.
To be Published
|
|
2PTG
 
 | |
2PTH
 
 | | PEPTIDYL-TRNA HYDROLASE FROM ESCHERICHIA COLI | | Descriptor: | PEPTIDYL-TRNA HYDROLASE | | Authors: | Schmitt, E, Mechulam, Y, Fromant, M, Plateau, P, Blanquet, S. | | Deposit date: | 1997-03-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure at 1.2 A resolution and active site mapping of Escherichia coli peptidyl-tRNA hydrolase.
EMBO J., 16, 1997
|
|
2PTK
 
 | | CHICKEN SRC TYROSINE KINASE | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Williams, J.C, Wierenga, R. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-24 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions
J.Mol.Biol., 274, 1997
|
|
2PTL
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN-BINDING DOMAIN OF PROTEIN L. COMPARISON WITH THE IGG-BINDING DOMAINS OF PROTEIN G | | Descriptor: | PROTEIN L | | Authors: | Wikstroem, M, Drakenberg, T, Forsen, S, Sjoebring, U, Bjoerck, L. | | Deposit date: | 1994-08-12 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G.
Biochemistry, 33, 1994
|
|
2PTM
 
 | | Structure and rearrangements in the carboxy-terminal region of SpIH channels | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, COBALT HEXAMMINE(III), Hyperpolarization-activated (Ih) channel | | Authors: | Flynn, G.E, Black, K.D, Islas, L.D, Sankaran, B, Zagotta, W.N. | | Deposit date: | 2007-05-08 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure and rearrangements in the carboxy-terminal region of SpIH channels.
Structure, 15, 2007
|
|
2PTN
 
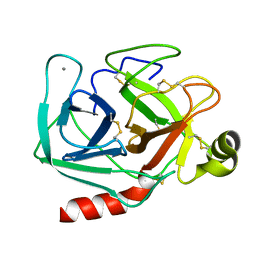 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, TRYPSIN | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
2PTQ
 
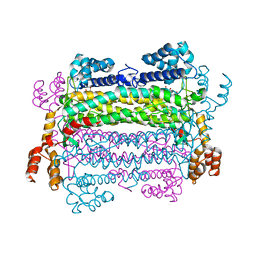 | |
2PTR
 
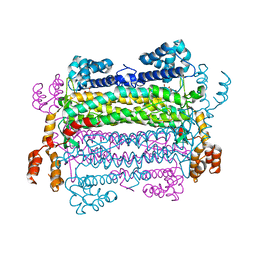 | |
2PTS
 
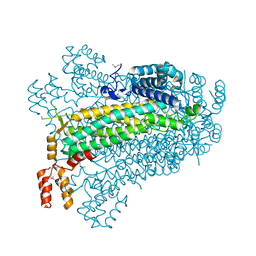 | |
2PTT
 
 | |
2PTU
 
 | | Structure of NK cell receptor 2B4 (CD244) | | Descriptor: | Natural killer cell receptor 2B4 | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
2PTV
 
 | | Structure of NK cell receptor ligand CD48 | | Descriptor: | CD48 antigen | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
2PTW
 
 | | Crystal Structure of the T. brucei enolase complexed with sulphate, identification of a metal binding site IV | | Descriptor: | 1,2-ETHANEDIOL, Enolase, SULFATE ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PTX
 
 | | Crystal Structure of the T. brucei enolase complexed with sulphate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, SULFATE ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PTY
 
 | | Crystal Structure of the T. brucei enolase complexed with PEP | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHOENOLPYRUVATE, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PTZ
 
 | | Crystal Structure of the T. brucei enolase complexed with phosphonoacetohydroxamate (PAH), His156-out conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHONOACETOHYDROXAMIC ACID, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PU0
 
 | | Crystal Structure of the T. brucei enolase complexed with phosphonoacetohydroxamate (PAH), His156-in conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHONOACETOHYDROXAMIC ACID, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PU1
 
 | | Crystal Structure of the T. brucei enolase complexed with Fluoro-phosphonoacetohydroxamate (FPAH) | | Descriptor: | 1,2-ETHANEDIOL, Enolase, ZINC ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PU2
 
 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | Descriptor: | 2-[(1R)-1-CARBOXY-2-(4-HYDROXYPHENYL)ETHYL]-1,3-DIOXOISOINDOLINE-5-CARBOXYLIC ACID, Beta-lactamase, PHOSPHATE ION | | Authors: | Babaoglu, K, Shoichet, B.K. | | Deposit date: | 2007-05-08 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
2PU3
 
 | | Structural adaptation of endonuclease I from the cold-adapted and halophilic bacterium Vibrio salmonicida | | Descriptor: | CHLORIDE ION, Endonuclease I, MAGNESIUM ION | | Authors: | Altermark, B, Helland, R, Moe, E, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2007-05-08 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural adaptation of endonuclease I from the cold-adapted and halophilic bacterium Vibrio salmonicida.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
