2MKB
 
 | |
1IIE
 
 | |
4M3I
 
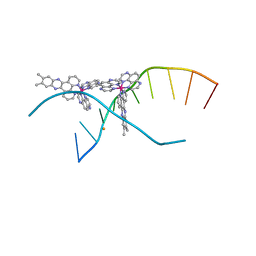 | | X-ray crystal structure of the ruthenium complex [Ru(TAP)2(dppz-{Me2})]2+ bound to d(CCGGTACCGG) | | Descriptor: | (11,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium(2+), BARIUM ION, Synthetic DNA CCGGTACCGG | | Authors: | Niyazi, H, Teixeira, S, Mitchell, E, Forsyth, T, Cardin, C. | | Deposit date: | 2013-08-06 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of the ruthenium complex [Ru(TAP)2(dppz-{Me2})]2+ bound to d(CCGGTACCGG)
To be Published
|
|
1IY4
 
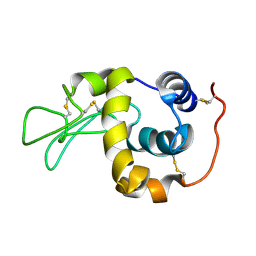 | | Solution structure of the human lysozyme at 35 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1BDY
 
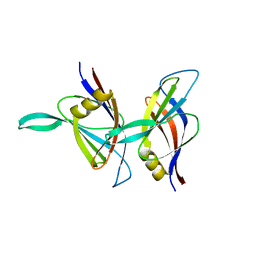 | | C2 DOMAIN FROM PROTEIN KINASE C DELTA | | Descriptor: | PROTEIN KINASE C | | Authors: | Pappa, H, Murray-Rust, J, Dekker, L.V, Parker, P.J, Mcdonald, N.Q. | | Deposit date: | 1998-05-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the C2 domain from protein kinase C-delta.
Structure, 6, 1998
|
|
2LZQ
 
 | |
2M21
 
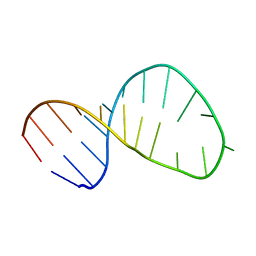 | | Solution structure of the Tetrahymena telomerase RNA stem IV terminal loop | | Descriptor: | 5'-R(*GP*GP*CP*GP*AP*UP*AP*CP*AP*CP*UP*AP*UP*UP*UP*AP*UP*CP*GP*CP*C)-3' | | Authors: | Cash, D.D, Richards, R.J, Wu, H, Trantirek, L, O'Connor, C.M, Feigon, J, Collins, K. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function.
Rna, 12, 2006
|
|
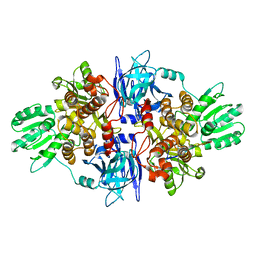
3O8C
 
 | |
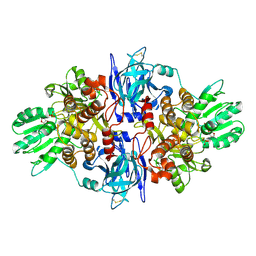
3O8D
 
 | |
1J6Q
 
 | | Solution structure and characterization of the heme chaperone CcmE | | Descriptor: | cytochrome c maturation protein E | | Authors: | Arnesano, F, Banci, L, Barker, P.D, Bertini, I, Rosato, A, Su, X.C, Viezzoli, M.S. | | Deposit date: | 2002-04-30 | | Release date: | 2002-12-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and characterization of the heme chaperone CcmE
Biochemistry, 41, 2002
|
|
3SVW
 
 | |
3SXT
 
 | |
4LJO
 
 | | Structure of an active ligase (HOIP)/ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, IMIDAZOLE, Polyubiquitin-C, ... | | Authors: | Rana, R.R, Stieglitz, B, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
2IWG
 
 | | COMPLEX BETWEEN THE PRYSPRY DOMAIN OF TRIM21 AND IGG FC | | Descriptor: | 52 KDA RO PROTEIN, IG GAMMA-1 CHAIN C, alpha-L-fucopyranose, ... | | Authors: | James, L.C, Keeble, A.H, Rhodes, D.A, Trowsdale, J. | | Deposit date: | 2006-06-30 | | Release date: | 2007-03-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Pryspry-Mediated Tripartite Motif (Trim) Protein Function.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1BY6
 
 | |
5QC3
 
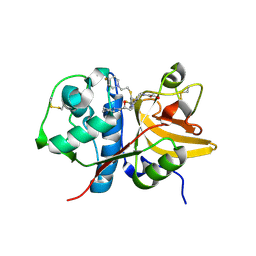 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-{3-[3-{[2-(4-fluoropiperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-[(2S)-2-hydroxy-3-(piperidin-1-yl)propyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}-2-hydroxyethan-1-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
3RN0
 
 | |
5QCB
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, tert-butyl 4-(2-{3-[3-{[(3-methylbut-2-enoyl)amino]methyl}-4-(trifluoromethyl)phenyl]-1-[3-(morpholin-4-yl)propyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}-2-oxoethyl)piperidine-1-carboxylate | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCC
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, tert-butyl 4-(2-{3-(4-chloro-3-iodophenyl)-1-[3-(morpholin-4-yl)propyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}-2-oxoethyl)piperidine-1-carboxylate | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QC8
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-benzyl-1-{3-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}methanamine | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
4LJP
 
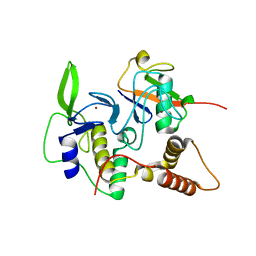 | | Structure of an active ligase (HOIP-H889A)/ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Polyubiquitin-C, ZINC ION | | Authors: | Rana, R.R, Stieglitz, B, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
5QCD
 
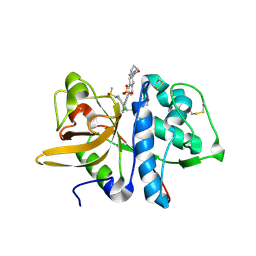 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, SULFATE ION, {(3S)-7-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]-1,2,3,4-tetrahydroisoquinolin-3-yl}(piperidin-1-yl)methanone | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
3O8R
 
 | |
2N79
 
 | | The structural and functional effects of the Familial Hypertrophic Cardiomyopathy-linked cardiac troponin C mutation, L29Q | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Robertson, I.M, Sevrieva, I, Li, M.X, Irving, M, Sun, Y, Sykes, B. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural and functional effects of the familial hypertrophic cardiomyopathy-linked cardiac troponin C mutation, L29Q.
J.MOL.CELL.CARDIOL., 87, 2015
|
|
3SJL
 
 | |
