7VM0
 
 | | Crystal structure of YojK from B.subtilis in complex with UDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glycosyl transferase family 1, ... | | Authors: | Hou, X.D, Guo, B.D, Rao, Y.J. | | Deposit date: | 2021-10-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly efficient production of rebaudioside D enabled by structure-guided engineering of bacterial glycosyltransferase YojK.
Front Bioeng Biotechnol, 10, 2022
|
|
1MV1
 
 | | The Tandem, Sheared PA Pairs in 5'(rGGCPAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1JCK
 
 | |
2X2I
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
1D68
 
 | |
1D20
 
 | | SOLUTION STRUCTURE OF PHAGE LAMBDA HALF-OPERATOR DNA | | Descriptor: | DNA (5'-D(*CP*GP*GP*TP*GP*AP*TP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*CP*AP*CP*CP*G)-3') | | Authors: | Baleja, J.D, Sykes, B.D. | | Deposit date: | 1990-08-01 | | Release date: | 1991-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of phage lambda half-operator DNA by use of NMR, restrained molecular dynamics, and NOE-based refinement.
Biochemistry, 29, 1990
|
|
1Q15
 
 | | Carbapenam Synthetase | | Descriptor: | CarA | | Authors: | Miller, M.T, Gerratana, B, Stapon, A, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Carbapenam Synthetase (CarA)
J.Biol.Chem., 278, 2003
|
|
3O9D
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd19 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3LHJ
 
 | | Crystal Structure of p38a Mitogen-Activated Protein Kinase in Complex with a Pyrazolopyridinone Inhibitor. | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-3-[1-(2,4-difluorophenyl)-7-methyl-6-oxo-6,7-dihydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-4-methylbenzamide | | Authors: | Mohr, C, Jordan, S. | | Deposit date: | 2010-01-22 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Discovery and evaluation of 7-alkyl-1,5-bis-aryl-pyrazolopyridinones as highly potent, selective, and orally efficacious inhibitors of p38alpha mitogen-activated protein kinase.
J.Med.Chem., 53, 2010
|
|
4HA0
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant R230D with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
153D
 
 | | CRYSTAL STRUCTURE OF A MISPAIRED DODECAMER, D(CGAGAATTC(O6ME)GCG)2, CONTAINING A CARCINOGENIC O6-METHYLGUANINE | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*AP*AP*TP*TP*CP*(6OG)P*CP*G)-3') | | Authors: | Ginell, S.L, Vojtechovsky, J, Gaffney, B, Jones, R, Berman, H.M. | | Deposit date: | 1993-12-16 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a mispaired dodecamer, d(CGAGAATTC(O6Me)GCG)2, containing a carcinogenic O6-methylguanine
Biochemistry, 33, 1994
|
|
1AGH
 
 | |
2C8R
 
 | | insuline(60sec) and UV laser excited fluorescence | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4H9U
 
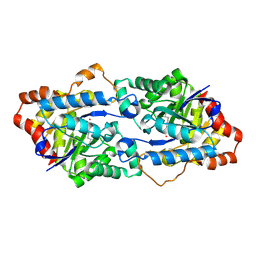 | | Structure of Geobacillus kaustophilus lactonase, wild-type with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
1UV6
 
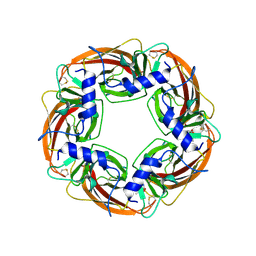 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with carbamylcholine | | Descriptor: | 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
4AEO
 
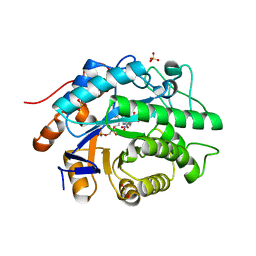 | | Structure of Xenobiotic Reductase B from Pseudomonas putida in complex with TNT | | Descriptor: | 2,4,6-TRINITROTOLUENE, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Carvalho, A.L, Mukhopadhyay, A, Romao, M.J, Bursakov, S, Kladova, A, Ramos, J.L, van Dillewijn, P. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of Aromatic Ring Reduction in the Xenobiotic Reductase B from Pseudomonas Putida
To be Published
|
|
476D
 
 | | CALCIUM FORM OF B-DNA UNDECAMER GCGAATTCGCG | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', CALCIUM ION | | Authors: | Minasov, G, Tereshko, V, Egli, M. | | Deposit date: | 1999-06-25 | | Release date: | 1999-07-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-resolution crystal structures of B-DNA reveal specific influences of divalent metal ions on conformation and packing.
J.Mol.Biol., 291, 1999
|
|
2C8Q
 
 | | insuline(1sec) and UV laser excited fluorescence | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
478D
 
 | |
1KJI
 
 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-AMPPCP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
3QD5
 
 | |
8IGC
 
 | | Crystal structure of Bak bound to Bnip5 BH3 | | Descriptor: | Bcl-2 homologous antagonist/killer, Protein BNIP5 | | Authors: | Ku, B, Lim, D. | | Deposit date: | 2023-02-20 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Crystal structure of Bak bound to the BH3 domain of Bnip5, a noncanonical BH3 domain-containing protein.
Proteins, 92, 2024
|
|
1KJ9
 
 | | Crystal structure of purt-encoded glycinamide ribonucleotide transformylase complexed with Mg-ATP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
4P1U
 
 | |
3HKY
 
 | | HCV NS5B polymerase genotype 1b in complex with 1,5 benzodiazepine 6 | | Descriptor: | (11S)-10-[(2,5-dimethyl-1,3-oxazol-4-yl)carbonyl]-11-{2-fluoro-4-[(2-methylprop-2-en-1-yl)oxy]phenyl}-3,3-dimethyl-2,3,4,5,10,11-hexahydrothiopyrano[3,2-b][1,5]benzodiazepin-6-ol 1,1-dioxide, CHLORIDE ION, RNA-directed RNA polymerase, ... | | Authors: | Nyanguile, O, De Bondt, H.L. | | Deposit date: | 2009-05-26 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus
J.Virol., 84, 2010
|
|
