1MEK
 
 | | HUMAN PROTEIN DISULFIDE ISOMERASE, NMR, 40 STRUCTURES | | Descriptor: | PROTEIN DISULFIDE ISOMERASE | | Authors: | Kemmink, J, Darby, N.J, Dijkstra, K, Nilges, M, Creighton, T.E. | | Deposit date: | 1996-04-16 | | Release date: | 1997-04-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the N-terminal thioredoxin-like domain of protein disulfide isomerase using multidimensional heteronuclear 13C/15N NMR spectroscopy.
Biochemistry, 35, 1996
|
|
2ROY
 
 | | TRANSTHYRETIN (ALSO CALLED PREALBUMIN) COMPLEX WITH 3',5'-DINITRO-N-ACETYL-L-THYRONINE | | Descriptor: | 3',5'-DINITRO-N-ACETYL-L-THYRONINE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 1996-10-23 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of human transthyretin complexed with thyroxine at 2.0 A resolution and 3',5'-dinitro-N-acetyl-L-thyronine at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1IET
 
 | |
1IEU
 
 | |
2CBP
 
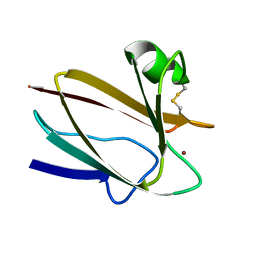 | | CUCUMBER BASIC PROTEIN, A BLUE COPPER PROTEIN | | Descriptor: | COPPER (II) ION, CUCUMBER BASIC PROTEIN | | Authors: | Guss, J.M, Freeman, H.C. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a phytocyanin, the basic blue protein from cucumber, refined at 1.8 A resolution.
J.Mol.Biol., 262, 1996
|
|
2SOC
 
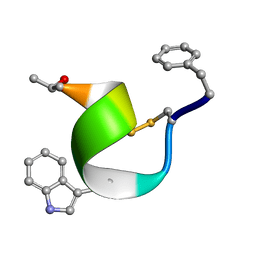 | |
2TRH
 
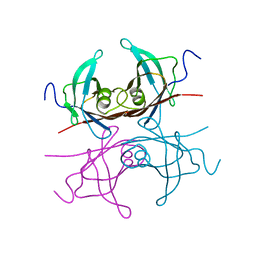 | |
2TRY
 
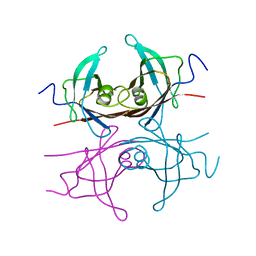 | |
2ROX
 
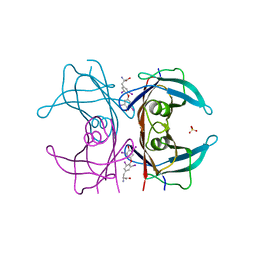 | | TRANSTHYRETIN (ALSO CALLED PREALBUMIN) COMPLEX WITH THYROXINE (T4) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SULFATE ION, TRANSTHYRETIN | | Authors: | Wojtczak, A, Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 1996-10-23 | | Release date: | 1997-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of human transthyretin complexed with thyroxine at 2.0 A resolution and 3',5'-dinitro-N-acetyl-L-thyronine at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1PEN
 
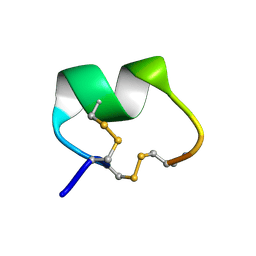 | | ALPHA-CONOTOXIN PNI1 | | Descriptor: | ALPHA-CONOTOXIN PNIA | | Authors: | Hu, S.-H, Gehrmann, J, Guddat, L.W, Alewood, P.F, Craik, D.J, Martin, J.L. | | Deposit date: | 1996-01-29 | | Release date: | 1997-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A crystal structure of the neuronal acetylcholine receptor antagonist, alpha-conotoxin PnIA from Conus pennaceus.
Structure, 4, 1996
|
|
6UPJ
 
 | | HIV-2 PROTEASE/U99294 COMPLEX | | Descriptor: | 6,7,8,9-TETRAHYDRO-4-HYDROXY-3-(1-PHENYLPROPYL)CYCLOHEPTA[B]PYRAN-2-ONE, HIV-2 PROTEASE | | Authors: | Watenpaugh, K.D, Mulichak, A.M, Finzel, B.C. | | Deposit date: | 1996-12-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Use of medium-sized cycloalkyl rings to enhance secondary binding: discovery of a new class of human immunodeficiency virus (HIV) protease inhibitors.
J.Med.Chem., 38, 1995
|
|
1DEF
 
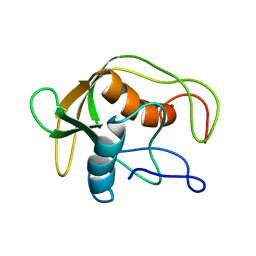 | |
1OIB
 
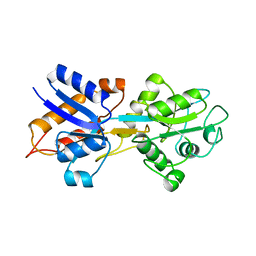 | |
1OLD
 
 | |
1ZAP
 
 | | SECRETED ASPARTIC PROTEASE FROM C. ALBICANS | | Descriptor: | N-ethyl-N-[(4-methylpiperazin-1-yl)carbonyl]-D-phenylalanyl-N-[(1S,2S,4R)-4-(butylcarbamoyl)-1-(cyclohexylmethyl)-2-hydroxy-5-methylhexyl]-L-norleucinamide, SECRETED ASPARTIC PROTEINASE, ZINC ION | | Authors: | Abad-Zapatero, C, Muchmore, S.W. | | Deposit date: | 1996-01-16 | | Release date: | 1997-04-21 | | Last modified: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a secreted aspartic protease from C. albicans complexed with a potent inhibitor: implications for the design of antifungal agents.
Protein Sci., 5, 1996
|
|
1JUG
 
 | | LYSOZYME FROM ECHIDNA MILK (TACHYGLOSSUS ACULEATUS) | | Descriptor: | CALCIUM ION, LYSOZYME | | Authors: | Guss, J.M. | | Deposit date: | 1996-10-13 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the calcium-binding echidna milk lysozyme at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1SOC
 
 | |
1VLS
 
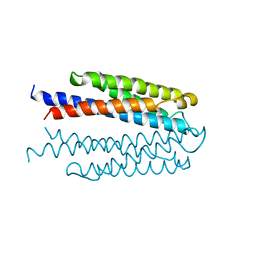 | | LIGAND BINDING DOMAIN OF THE WILD-TYPE ASPARTATE RECEPTOR | | Descriptor: | ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Yeh, J.I, Biemann, H.-P, Prive, G, Pandit, J, Koshland Junior, D.E. | | Deposit date: | 1996-09-17 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor.
J.Mol.Biol., 262, 1996
|
|
1RT2
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
1AAY
 
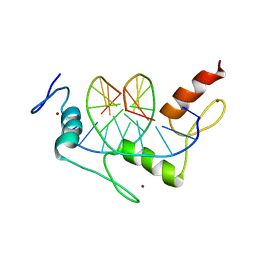 | | ZIF268 ZINC FINGER-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Rould, M.A, Pabo, C.O. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions.
Structure, 4, 1996
|
|
1ZDI
 
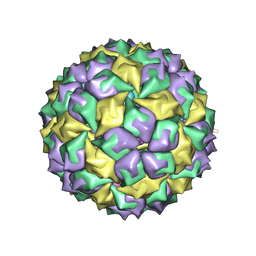 | | RNA BACTERIOPHAGE MS2 COAT PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (RNA BACTERIOPHAGE MS2 COAT PROTEIN), RNA (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*U P*GP*U)-3') | | Authors: | Valegard, K, Van Den Worm, S, Liljas, L. | | Deposit date: | 1996-09-24 | | Release date: | 1997-04-21 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions.
J.Mol.Biol., 270, 1997
|
|
1OSP
 
 | |
1KDN
 
 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Cherfils, J, Xu, Y.W, Morera, S, Janin, J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1KCE
 
 | | E. COLI THYMIDYLATE SYNTHASE MUTANT E58Q IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Rutenber, E.E, Stout, T.J, Stroud, R.M. | | Deposit date: | 1996-10-22 | | Release date: | 1997-04-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An essential role for water in an enzyme reaction mechanism: the crystal structure of the thymidylate synthase mutant E58Q.
Biochemistry, 35, 1996
|
|
1LDY
 
 | |
