1GWA
 
 | | Triiodide derivative of porcine pancreas elastase | | Descriptor: | CALCIUM ION, ELASTASE 1, IODIDE ION, ... | | Authors: | Evans, G, Bricogne, G. | | Deposit date: | 2002-03-13 | | Release date: | 2002-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Triiodide Derivatization and Combinatorial Counter-Ion Replacement: Two Methods for Enhancing Phasing Signal Using Laboratory Cu Kalpha X-Ray Equipment
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HO2
 
 | | NMR STRUCTURE OF THE POTASSIUM CHANNEL FRAGMENT L45 IN MICELLES | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN | | Authors: | Ohlenschlager, O, Hojo, H, Ramachandran, R, Gorlach, M, Haris, P.I. | | Deposit date: | 2000-12-08 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the S4-S5 segment of the Shaker potassium channel.
Biophys.J., 82, 2002
|
|
1HQ8
 
 | | CRYSTAL STRUCTURE OF THE MURINE NK CELL-ACTIVATING RECEPTOR NKG2D AT 1.95 A | | Descriptor: | NKG2-D | | Authors: | Wolan, D.W, Teyton, L, Rudolph, M.G, Villmow, B, Bauer, S, Busch, D.H, Wilson, I.A. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A.
Nat.Immunol., 2, 2001
|
|
9FO8
 
 | | 5_SL5b_GC of the 5_SL5 RNA | | Descriptor: | RNA (29-MER) | | Authors: | Mertinkus, K.R, Oxenfarth, A, Schwalbe, H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Dissecting the Conformational Heterogeneity of Stem-Loop Substructures of the Fifth Element in the 5'-Untranslated Region of SARS-CoV-2.
J.Am.Chem.Soc., 2024
|
|
9FO9
 
 | |
1GXN
 
 | | Family 10 polysaccharide lyase from Cellvibrio cellulosa | | Descriptor: | PECTATE LYASE | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1J8E
 
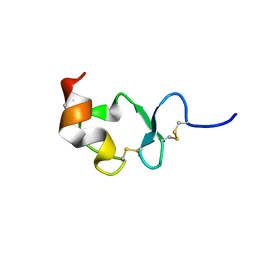 | | Crystal structure of ligand-binding repeat CR7 from LRP | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN 1 | | Authors: | Simonovic, M, Dolmer, K, Huang, W, Strickland, D.K, Volz, K, Gettins, P.G.W. | | Deposit date: | 2001-05-21 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor.
Biochemistry, 40, 2001
|
|
1IYG
 
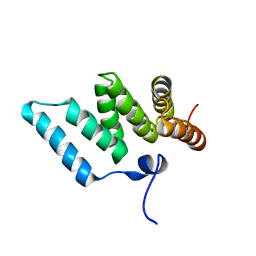 | | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA | | Descriptor: | Hypothetical protein (2010003O14) | | Authors: | Ohashi, W, Hirota, H, Yamazaki, T, Koshiba, S, Hamada, T, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-14 | | Release date: | 2003-02-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA
To be Published
|
|
1J85
 
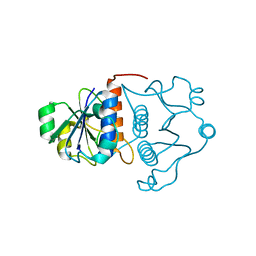 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
1IGQ
 
 | |
1B9K
 
 | |
1BDW
 
 | |
1IMW
 
 | | Peptide Antagonist of IGFBP-1 | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1J8T
 
 | | Catalytic Domain of Human Phenylalanine Hydroxylase Fe(II) | | Descriptor: | FE (II) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Andersen, O.A, Flatmark, T, Hough, E. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin.
J.Mol.Biol., 314, 2001
|
|
1IU1
 
 | | Crystal structure of human gamma1-adaptin ear domain | | Descriptor: | gamma1-adaptin | | Authors: | Nogi, T, Shiba, Y, Kawasaki, M, Shiba, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Takatsu, H, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2002-02-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain.
Nat.Struct.Biol., 9, 2002
|
|
1IH8
 
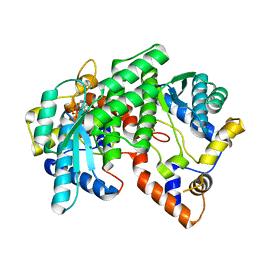 | | NH3-dependent NAD+ Synthetase from Bacillus subtilis Complexed with AMP-CPP and Mg2+ ions. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) synthetase | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, DeLucas, L. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1BFV
 
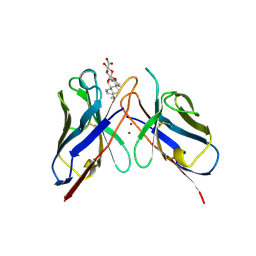 | | MONOCLONAL ANTIBODY FRAGMENT FV4155 FROM E. COLI | | Descriptor: | ESTRIOL 3-(B-D-GLUCURONIDE), FV4155, ZINC ION | | Authors: | Trinh, C.H, Phillips, S.E.V. | | Deposit date: | 1997-05-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibody fragment Fv4155 bound to two closely related steroid hormones: the structural basis of fine specificity.
Structure, 5, 1997
|
|
1BG2
 
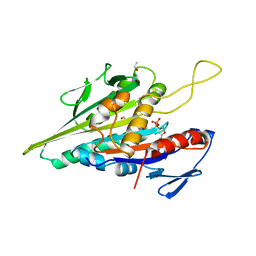 | | HUMAN UBIQUITOUS KINESIN MOTOR DOMAIN | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, KINESIN, ... | | Authors: | Kull, F.J, Sablin, E.P, Lau, R, Fletterick, R.J, Vale, R.D. | | Deposit date: | 1998-06-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the kinesin motor domain reveals a structural similarity to myosin.
Nature, 380, 1996
|
|
1I8A
 
 | | FAMILY 9 CARBOHYDRATE-BINDING MODULE FROM THERMOTOGA MARITIMA XYLANASE 10A WITH GLUCOSE | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Warren, R.A.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-03-12 | | Release date: | 2001-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms.
Biochemistry, 40, 2001
|
|
1I9C
 
 | |
1IA8
 
 | | THE 1.7 A CRYSTAL STRUCTURE OF HUMAN CELL CYCLE CHECKPOINT KINASE CHK1 | | Descriptor: | CHK1 CHECKPOINT KINASE, SULFATE ION | | Authors: | Chen, P, Luo, C, Deng, Y, Ryan, K, Register, J, Margosiak, S, Tempczyk-Russell, A, Nguyen, B, Myers, P, Lundgren, K, Chen Kan, C.-C, O'Connor, P.M. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation.
Cell(Cambridge,Mass.), 100, 2000
|
|
9GL0
 
 | | Crystal Structure of Acetylpolyamine aminohydrolase (ApaH) from Legionella pneumophila | | Descriptor: | Acetylpolyamine aminohydrolase, POTASSIUM ION, ZINC ION | | Authors: | Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria
Nature Communications, 2024
|
|
9GL1
 
 | | Crystal Structure of Acetylpolyamine aminohydrolase (ApaH) from Legionella cherrii | | Descriptor: | Acetylpolyamine aminohydrolase, POTASSIUM ION, ZINC ION | | Authors: | Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria
Nature Communications, 2024
|
|
8ZNS
 
 | |
1HXS
 
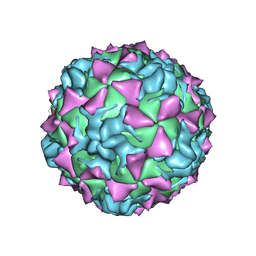 | | CRYSTAL STRUCTURE OF MAHONEY STRAIN OF POLIOVIRUS AT 2.2A RESOLUTION | | Descriptor: | GENOME POLYPROTEIN, COAT PROTEIN VP1, COAT PROTEIN VP2, ... | | Authors: | Miller, S.T, Hogle, J.M, Filman, D.J. | | Deposit date: | 2001-01-16 | | Release date: | 2002-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ab initio phasing of high-symmetry macromolecular complexes: successful phasing of authentic poliovirus data to 3.0 A resolution.
J.Mol.Biol., 307, 2001
|
|
