8BGW
 
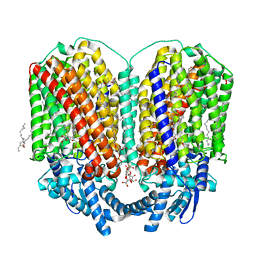 | | CryoEM structure of quinol-dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans at 2.2 A resolution | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, FE (III) ION, ... | | Authors: | Flynn, A, Antonyuk, S.V, Eady, R.R, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2022-10-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | A 2.2 angstrom cryoEM structure of a quinol-dependent NO Reductase shows close similarity to respiratory oxidases.
Nat Commun, 14, 2023
|
|
8BMY
 
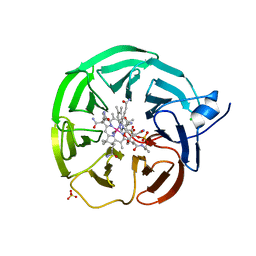 | | Bacteroides thetaiotaomicron surface lipoprotein bound to cyanocobalamin | | Descriptor: | CHLORIDE ION, CYANOCOBALAMIN, Putative surface layer protein, ... | | Authors: | Abellon-Ruiz, J, Jana, K, Silale, A, Basle, A, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2022-11-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
8BLW
 
 | |
8BN0
 
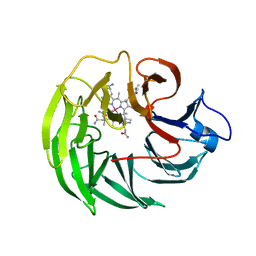 | | Bacteroides thetaiotaomicron surface protein BT1954 bound to | | Descriptor: | CHLORIDE ION, COB(II)INAMIDE, CYANIDE ION, ... | | Authors: | Abellon-Ruiz, J, Jana, K, Silale, A, Basle, A, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2022-11-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
6MK4
 
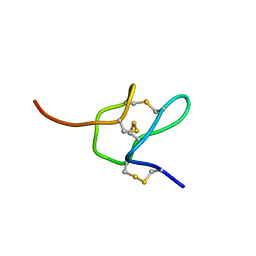 | |
6MXY
 
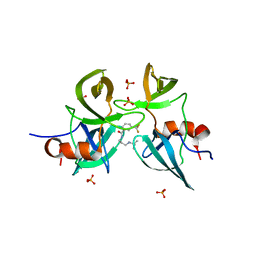 | |
8BWW
 
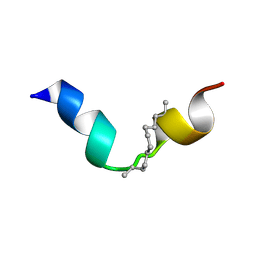 | |
6MJJ
 
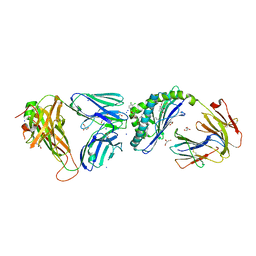 | | Crystal structure of the mCD1d/xxm (JJ290) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
8AHQ
 
 | | VirD/holo-ACP5b of Streptomyces virginiae complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, CHLORIDE ION, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8AHZ
 
 | | Native VirD of Streptomyces virginiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enoyl-CoA hydratase, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8B01
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc | | Descriptor: | DECANE, DOCOSANE, HEXANE, ... | | Authors: | Davies, J.S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2022-09-06 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
8AAC
 
 | |
8AIG
 
 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8ALL
 
 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
6MWW
 
 | | LasR LBD:BB0126 complex | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1R,3R,5R)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
8AGG
 
 | |
8B4S
 
 | | Antimicrobial peptide capitellacin from polychaeta Capitella teleta in DPC (dodecylphosphocholine) micelles, dimeric form | | Descriptor: | BRICHOS domain-containing protein | | Authors: | Mironov, P.A, Reznikova, O.V, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Dimerization of the beta-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study.
Biomolecules, 14, 2024
|
|
8B9S
 
 | |
8BOB
 
 | | Structural basis for negative regulation of the maltose system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HTH-type transcriptional regulator MalT, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Chai, J, Wu, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for negative regulation of the Escherichia coli maltose system.
Nat Commun, 14, 2023
|
|
8BL3
 
 | |
8BL6
 
 | |
6N7Q
 
 | |
6MXZ
 
 | | Structure of 53BP1 Tudor domains in complex with small molecule UNC3474 | | Descriptor: | FORMIC ACID, N-[3-(tert-butylamino)propyl]-3-(propan-2-yl)benzamide, TP53-binding protein 1 | | Authors: | Cui, G, Botuyan, M.V, Schuller, D.J, Mer, G. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering.
Nat Commun, 14, 2023
|
|
6MRT
 
 | | 12-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
8A7O
 
 | |
