6QSP
 
 | | Ketosynthase (ApeO) in Complex with its Chain Length Factor (ApeC) from Xenorhabdus doucetiae | | Descriptor: | Beta-ketoacyl synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grammbitter, G.L.C, Schmalhofer, M, Groll, M, Bode, H. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An Uncommon Type II PKS Catalyzes Biosynthesis of Aryl Polyene Pigments.
J.Am.Chem.Soc., 141, 2019
|
|
8IN9
 
 | | The structure of the GfsA KSQ-AT didomain in complex with the GfsA ACP domain | | Descriptor: | N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, Polyketide synthase | | Authors: | Chisuga, T, Murakami, S, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-Based Analysis of Transient Interactions between Ketosynthase-like Decarboxylase and Acyl Carrier Protein in a Loading Module of Modular Polyketide Synthase.
Acs Chem.Biol., 18, 2023
|
|
4Z8D
 
 | |
7VEE
 
 | | The ligand-free structure of GfsA KSQ-AT didomain | | Descriptor: | GLYCEROL, Polyketide synthase | | Authors: | Chisuga, T, Miyanaga, A, Nagai, A, Kudo, F, Eguchi, T. | | Deposit date: | 2021-09-08 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight into the Reaction Mechanism of Ketosynthase-Like Decarboxylase in a Loading Module of Modular Polyketide Synthases.
Acs Chem.Biol., 17, 2022
|
|
7VEF
 
 | | The structure of GfsA KSQ-AT didomain in complex with a malonate substrate analog | | Descriptor: | GLYCEROL, N-(2-acetamidoethyl)-2-nitro-ethanamide, Polyketide synthase | | Authors: | Chisuga, T, Miyanaga, A, Nagai, A, Kudo, F, Eguchi, T. | | Deposit date: | 2021-09-08 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insight into the Reaction Mechanism of Ketosynthase-Like Decarboxylase in a Loading Module of Modular Polyketide Synthases.
Acs Chem.Biol., 17, 2022
|
|
5D6N
 
 | | Crystal structure of a mycobacterial protein | | Descriptor: | Acyl-CoA synthase | | Authors: | Li, W.J, Bi, L.J. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of FadD32, an enzyme essential for mycolic acid biosynthesis in mycobacteria.
Sci Rep, 5, 2015
|
|
3FK5
 
 | |
3T5Y
 
 | | Crystal structure of CerJ from Streptomyces tendae - malonic acid covalently linked to the catalytic Cystein C116 | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
7CPX
 
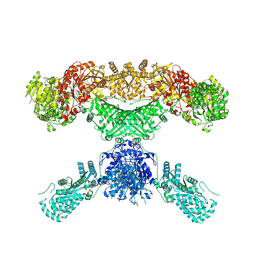 | | Lovastatin nonaketide synthase | | Descriptor: | Lovastatin nonaketide synthase, polyketide synthase component, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2020-08-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
7ZSK
 
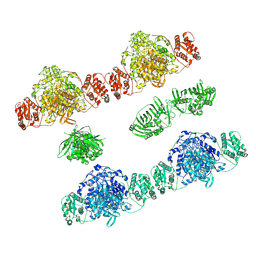 | |
8OTM
 
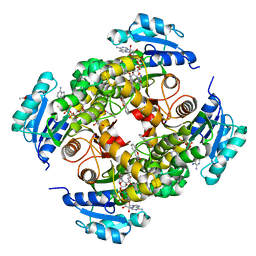 | | structure of InhA from mycobacterium tuberculosis in complex with N-((1-(3-hydroxy-4-phenoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl)-2-oxo-2H-chromene-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-oxidanylidene-~{N}-[[1-[(3-oxidanyl-4-phenoxy-phenyl)methyl]-1,2,3-triazol-4-yl]methyl]chromene-3-carboxamide, ACETATE ION, ... | | Authors: | Chebaiki, M, Maveyraud, L, Tamhaev, R, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA.
Eur.J.Med.Chem., 259, 2023
|
|
8OTN
 
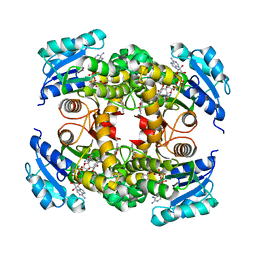 | | structure of InhA from mycobacterium tuberculosis in complex with inhibitor 7-((1-(3-Hydroxy-4-phenoxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-4-methyl-2H-chromen-2-one | | Descriptor: | 4-methyl-7-[[1-[(3-oxidanyl-4-phenoxy-phenyl)methyl]-1,2,3-triazol-4-yl]methoxy]chromen-2-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chebaiki, M, Maveyraud, L, Tamhaev, R, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA.
Eur.J.Med.Chem., 259, 2023
|
|
2JBZ
 
 | | Crystal structure of the Streptomyces coelicolor holo-[Acyl-carrier-protein] Synthase (AcpS) in complex with coenzyme A at 1.6 A | | Descriptor: | COENZYME A, HOLO-[ACYL-CARRIER-PROTEIN] SYNTHASE, MAGNESIUM ION, ... | | Authors: | Dall'Aglio, P, Arthur, C, Law, C.K.E, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
4DXE
 
 | | 2.52 Angstrom resolution crystal structure of the acyl-carrier-protein synthase (AcpS)-acyl carrier protein (ACP) protein-protein complex from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Acyl carrier protein, MALONATE ION, acyl-carrier-protein synthase | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-27 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | 2.52 Angstrom resolution crystal structure of the acyl-carrier-protein synthase (AcpS)-acyl carrier protein (ACP) protein-protein complex from Staphylococcus aureus subsp. aureus COL
To be Published
|
|
7S2X
 
 | | Structure of SalC, a gamma-lactam-beta-lactone bicyclase for salinosporamide biosynthesis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, POTASSIUM ION, SalC | | Authors: | Chen, P.Y.-T, Trivella, D.B.B, Bauman, K.D, Moore, B.S. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Enzymatic assembly of the salinosporamide gamma-lactam-beta-lactone anticancer warhead.
Nat.Chem.Biol., 18, 2022
|
|
5BY7
 
 | | AbyA1 - tetronic acid condensing enzyme | | Descriptor: | 3-oxoacyl-ACP synthase III | | Authors: | Byrne, M.J, Race, P.R. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of AbyA1, a tetronic acid condensing enzyme from the Abyssomicin polyketide synthase.
To Be Published
|
|
8HTA
 
 | |
5XUM
 
 | | Crystal structure of Thermotoga maritima holo-[acyl-carrier-protein] synthase (AcpS) | | Descriptor: | GLYCEROL, Holo-[acyl-carrier-protein] synthase | | Authors: | Liao, Y.P, Wang, D.L, Yin, D.P, Zhang, Q.Y, Wang, Y.M, Wang, D.Q, Zhu, H.X, Chen, S. | | Deposit date: | 2017-06-23 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of acyl carrier protein synthases (AcpS) from three Gram-negative bacteria
To Be Published
|
|
6FIK
 
 | | ACP2 crosslinked to the KS of the loading/condensing region of the CTB1 PKS | | Descriptor: | Polyketide synthase | | Authors: | Herbst, D.A, Huitt-Roehl, C.R, Jakob, R.P, Townsend, C.A, Maier, T. | | Deposit date: | 2018-01-18 | | Release date: | 2018-03-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | The structural organization of substrate loading in iterative polyketide synthases.
Nat. Chem. Biol., 14, 2018
|
|
5XUK
 
 | | Crystal structure of Helicobacter pylori holo-[acyl-carrier-protein] synthase (AcpS) in complex with coenzyme A | | Descriptor: | COENZYME A, Holo-[acyl-carrier-protein] synthase | | Authors: | Liao, Y.P, Wang, D.L, Yin, D.P, Zhang, Q.Y, Wang, Y.M, Wang, D.Q, Zhu, H.X, Chen, S. | | Deposit date: | 2017-06-23 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of acyl carrier protein synthases (AcpS) from three Gram-negative bacteria
To Be Published
|
|
5XU7
 
 | | Crystal structure of Escherichia coli holo-[acyl-carrier-protein] synthase (AcpS) | | Descriptor: | CHLORIDE ION, GLYCEROL, Holo-[acyl-carrier-protein] synthase | | Authors: | Liao, Y.P, Wang, D.L, Yin, D.P, Zhang, Q.Y, Wang, Y.M, Wang, D.Q, Zhu, H.X, Chen, S. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of acyl carrier protein synthases (AcpS) from three Gram-negative bacteria
To Be Published
|
|
5XUH
 
 | | Crystal structure of Escherichia coli holo-[acyl-carrier-protein] synthase (AcpS) D9A mutant in complex with CoA | | Descriptor: | CHLORIDE ION, COENZYME A, GLYCEROL, ... | | Authors: | Liao, Y.P, Wang, D.L, Yin, D.P, Zhang, Q.Y, Wang, Y.M, Wang, D.Q, Zhu, H.X, Chen, S. | | Deposit date: | 2017-06-23 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of acyl carrier protein synthases (AcpS) from three Gram-negative bacteria
To Be Published
|
|
8H6S
 
 | | Structure of acyltransferase VinK in complex with the loading acyl carrier protein of vicenistatin PKS | | Descriptor: | MAGNESIUM ION, Malonyl-CoA-[acyl-carrier-protein] transacylase, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Kawada, K, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Transient Interactions of Acyltransferase VinK with the Loading Acyl Carrier Protein of the Vicenistatin Modular Polyketide Synthase.
Biochemistry, 62, 2023
|
|
5MY2
 
 | |
7TUI
 
 | |
