9I98
 
 | | Cryo-EM structure of Shigella flexneri LptDE bound by a Bicyclic peptide molecule (Compound 13) | | Descriptor: | 1-[3,5-bis(2-chloranylethanoyl)-1,3,5-triazinan-1-yl]-2-chloranyl-ethanone, Compound 13 - Bicyclic Peptide binder, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Allyjaun, S, Newman, H, Dunbar, E, Hardwick, S.W, Chirgadze, D.Y, van den Berg, B, Hubbard, J. | | Deposit date: | 2025-02-06 | | Release date: | 2025-10-15 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | High-Throughput Identification and Characterization of LptDE-Binding Bicycle Peptides Using Phage Display and Cryo-EM.
J.Med.Chem., 68, 2025
|
|
8JU9
 
 | |
9I7Z
 
 | | LecA in complex with 2-fluoro non-carbohydrate glycomimetic | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethanol, ACETATE ION, ... | | Authors: | Varrot, A. | | Deposit date: | 2025-02-03 | | Release date: | 2025-10-22 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Parkinson's Disease Drug Tolcapone and Analogues are Potent Glycomimetic Lectin Inhibitors of Pseudomonas aeruginosa LecA.
Angew.Chem.Int.Ed.Engl., 2025
|
|
4USD
 
 | | Human STK10 (LOK) with SB-633825 | | Descriptor: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
7Z3L
 
 | | Autotaxin in complex with hybrid compound ziritaxestat (GLPG1690) | | Descriptor: | 2-[[2-ethyl-8-methyl-6-[4-[2-(3-oxidanylazetidin-1-yl)-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Ford, P, Heckmann, B, Perrakis, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autotaxin facilitates selective LPA receptor signaling.
Cell Chem Biol, 30, 2023
|
|
8JT1
 
 | | COLLAGENASE FROM GRIMONTIA (VIBRIO) HOLLISAE 1706B COMPLEXED WITH GLY-PRO-HYP-GLY-PRO-HYP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-mer peptide, ... | | Authors: | Ueshima, S, Yaskawa, K, Takita, T, Mikami, B. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the catalytic mechanism of Grimontia hollisae collagenase through structural and mutational analyses.
Febs Lett., 597, 2023
|
|
8K8J
 
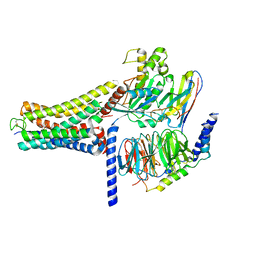 | | Cannabinoid Receptor 1 bound to Fenofibrate coupling MiniGsq and Nb35 Complex | | Descriptor: | Fenofibrate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tang, W.Q, Wang, T.X, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Fenofibrate Recognition and G q Protein Coupling Mechanisms of the Human Cannabinoid Receptor CB1.
Adv Sci, 11, 2024
|
|
4WHZ
 
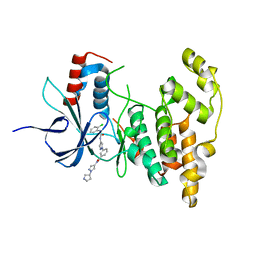 | |
9HUB
 
 | | D11 mAbs bound to alpha-Bungarotoxin at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-bungarotoxin, heavy chain, ... | | Authors: | Wade, J.W, Bohn, M.F, Laustsen, A.H, Morth, J.P. | | Deposit date: | 2024-12-21 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Rational design of antibodies with pH-dependent antigen-binding properties using structural insights from broadly neutralizing antibodies against alpha-neurotoxins.
Mabs, 17, 2025
|
|
8JMZ
 
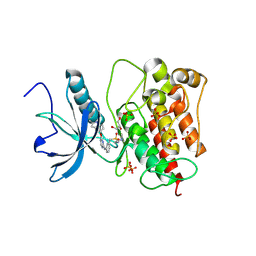 | | FGFR1 kinase domain with sulfatinib | | Descriptor: | Fibroblast growth factor receptor 1, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2023-06-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
4WS6
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-aminouracil, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7Z6T
 
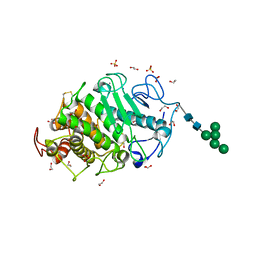 | | Aspergillus clavatus M36 protease without the propeptide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular metalloproteinase mep, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2022-03-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Aspergillus clavatus M36 protease without the propeptide
To Be Published
|
|
7SOD
 
 | |
7SOE
 
 | |
4USC
 
 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
7SOF
 
 | |
7T41
 
 | | Structure of MERS 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
8T1G
 
 | |
9J6O
 
 | | Crystal Structure of bromodomain of human CBP in complex with the inhibitor CZL-046 | | Descriptor: | (3~{S},5~{S})-1-[3,4-bis(fluoranyl)phenyl]-5-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]-3-fluoranyl-pyrrolidin-2-one, CHLORIDE ION, CREB-binding protein | | Authors: | Zhang, Y, Zhang, C, Chen, Z, Yang, H, Huang, X, Li, Y, Xu, Y. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of CZL-046 with an ( S )-3-Fluoropyrrolidin-2-one Scaffold as a p300 Bromodomain Inhibitor for the Treatment of Multiple Myeloma.
J.Med.Chem., 67, 2024
|
|
8JYY
 
 | |
9IXR
 
 | | Crystal structure of OXA-10 variant A124T in the complex with ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural insights into alterations in the substrate spectrum of serine-beta-lactamase OXA-10 from Pseudomonas aeruginosa by single amino acid substitutions.
Emerg Microbes Infect, 13, 2024
|
|
7SQ7
 
 | | Cryo-EM structure of mouse PI(3,5)P2-bound TRPML1 channel at 2.41 Angstrom resolution | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ON5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing nanobody Re5D06 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Nanobody Re5D06 | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
7SQ6
 
 | | Cryo-EM structure of mouse agonist ML-SA1-bound TRPML1 channel at 2.32 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4X46
 
 | | X-RAY structure thymidine phosphorylase from Salmonella typhimurium complex with SO4 at 2.19 A | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Thymidine phosphorylase | | Authors: | Balaev, V.V, Lashkov, A.A, Prokofev, I.I, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-RAY structure thymidine phosphorylase from Salmonella typhimurium complex with SO4 at 2.19 A
To Be Published
|
|
