5UVN
 
 | |
1N3W
 
 | | Engineered High-Affinity Maltose-Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2002-10-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
1N3X
 
 | | Ligand-free High-Affinity Maltose-Binding Protein | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2002-10-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
3HST
 
 | |
4EXK
 
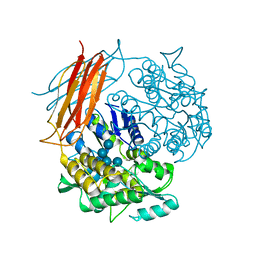 | | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 from Salmonella enterica | | Descriptor: | Maltose-binding periplasmic protein, uncharacterized protein chimera, TRIETHYLENE GLYCOL, ... | | Authors: | Nocek, B, Hatzos-Skintges, C, Jedrzejczak, R, Babnigg, G, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-30 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 form Salmonella enterica
To be Published
|
|
6XDS
 
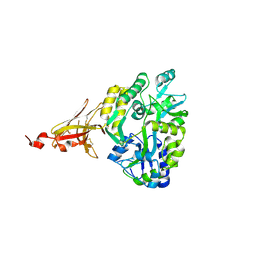 | | Crystal structure of MBP-TREM2 Ig domain fusion with fragment, 2-((4-bromophenyl)amino)ethan-1-ol | | Descriptor: | 2-[(4-bromophenyl)amino]ethan-1-ol, DIMETHYL SULFOXIDE, Sugar ABC transporter substrate-binding protein,Triggering receptor expressed on myeloid cells 2, ... | | Authors: | Su, H.P. | | Deposit date: | 2020-06-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | Development of a robust crystallization platform for immune receptor TREM2 using a crystallization chaperone strategy.
Protein Expr.Purif., 179, 2021
|
|
5V6Y
 
 | |
3LC8
 
 | |
6XRX
 
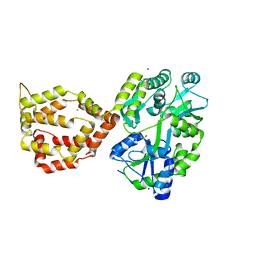 | | Crystal structure of the mosquito protein AZ1 as an MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Foo, A.C.Y. | | Deposit date: | 2020-07-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The mosquito protein AEG12 displays both cytolytic and antiviral properties via a common lipid transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3IOV
 
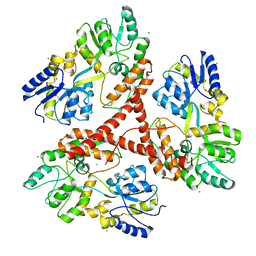 | |
4FEC
 
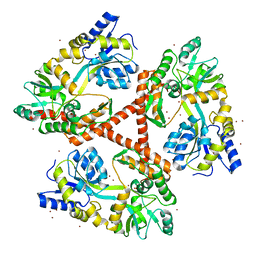 | | Crystal Structure of Htt36Q3H | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
5W0R
 
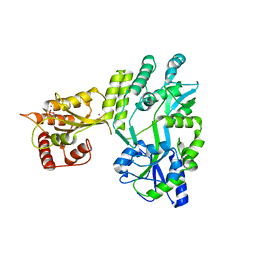 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cacodylic acid | | Descriptor: | CACODYLATE ION, CALCIUM ION, MBP fused activation-induced cytidine deaminase, ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
6XDJ
 
 | | Crystal Structure Analysis of MBP-SIN3 | | Descriptor: | Maltodextrin-binding protein,Transcriptional regulatory protein SIN3, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure Analysis of MBP-SIN3
To be published
|
|
3IO4
 
 | | Huntingtin amino-terminal region with 17 Gln residues - Crystal C90 | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin fusion protein, ZINC ION | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
4DXC
 
 | |
5W0U
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
1MH4
 
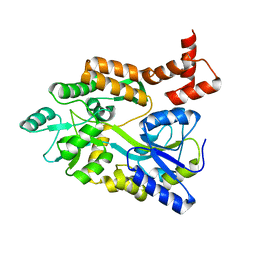 | | maltose binding-a1 homeodomain protein chimera, crystal form II | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
6YBK
 
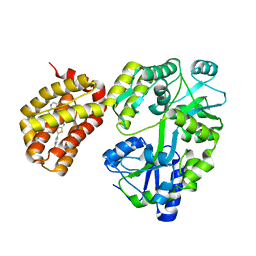 | | Structure of MBP-Mcl-1 in complex with compound 4d | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-(pyrazin-2-ylmethoxy)phenyl]propanoic acid, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
3LBS
 
 | |
1NL5
 
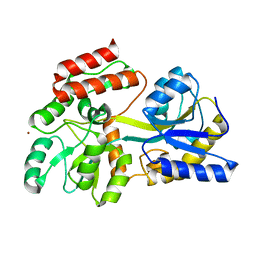 | | Engineered High-affinity Maltose-Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2003-01-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
6SJV
 
 | | Structure of HPV18 E6 oncoprotein in complex with mutant E6AP LxxLL motif | | Descriptor: | Maltodextrin-binding protein,Protein E6,Ubiquitin-protein ligase E3A, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Kostmann, C, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | Cellular target recognition by HPV18 and HPV49 oncoproteins
To be published
|
|
5W1C
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
3IO6
 
 | | Huntingtin amino-terminal region with 17 Gln residues - crystal C92-a | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein, HUNTINGTIN FUSION PROTEIN, ... | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
5WQ6
 
 | | Crystal Structure of hMNDA-PYD with MBP tag | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MBP tagged hMNDA-PYD, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
6SJA
 
 | | Structure of HPV16 E6 oncoprotein in complex with IRF3 LxxLL motif | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deciphering de molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
