1HP0
 
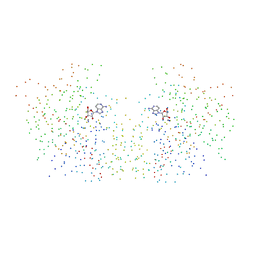 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX IN COMPLEX WITH THE SUBSTRATE ANALOGUE 3-DEAZA-ADENOSINE | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
5XDS
 
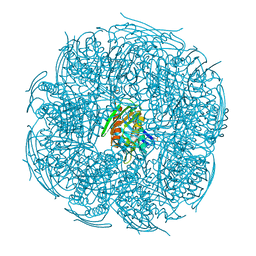 | | Crystal structure of Mycobacterium tuberculosis HisB bound with an inhibitor | | Descriptor: | (2S)-2-azanyl-3-(4H-1,2,4-triazol-3-yl)propanoic acid, CHLORIDE ION, Imidazoleglycerol-phosphate dehydratase, ... | | Authors: | Kumar, D, Jha, B, Ahangar, M.S, Kumar, B.B. | | Deposit date: | 2017-03-29 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of a triazole scaffold compound as an inhibitor of Mycobacterium tuberculosis imidazoleglycerol-phosphate dehydratase.
Proteins, 2021
|
|
5XW3
 
 | |
5XE4
 
 | |
5XZI
 
 | |
5XZJ
 
 | |
5YKA
 
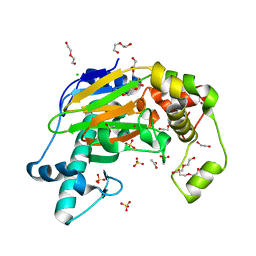 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with cobalt(II) | | Descriptor: | ACETATE ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-10-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
5YVZ
 
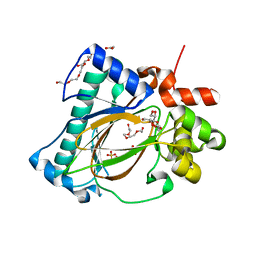 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with alphaketoglutarate and Fe(III) | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-11-28 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
5ZKK
 
 | |
5W99
 
 | | Pyridine synthase, PbtD, from GE2270 biosynthesis bound to TSP | | Descriptor: | 2,2'-(6-(2'-(aminomethyl)-[2,4'-bithiazol]-4-yl)pyridine-2,5-diyl)bis(thiazole-4-carboxylic acid), PbtD, SULFATE ION | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2017-06-22 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W98
 
 | | Pyridine synthase, PbtD, from GE2270 biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PbtD | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2017-06-22 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WRC
 
 | | Crystal structure of proteinase K from Engyodontium album | | Descriptor: | NITRATE ION, PRASEODYMIUM ION, Proteinase K | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Masuda, T, Inoue, S, Numata, K. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WR9
 
 | | Crystal structure of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WR8
 
 | | Thaumatin structure determined by SACLA at 1.55 Angstrom | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Sugahara, M. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5XEE
 
 | |
5WGB
 
 | |
5WJP
 
 | |
5Z1O
 
 | |
5YW0
 
 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with succinate and Fe(III) | | Descriptor: | ACETATE ION, FE (III) ION, GLYCEROL, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-11-28 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
5WRB
 
 | | Crystal structure of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WA3
 
 | |
5Z31
 
 | | LPS bound solution structure of WS2-KG18 | | Descriptor: | LYS-ASN-LYS-SER-ARG-VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
5WRA
 
 | | Crystal structure of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5ZCZ
 
 | | Solution structure of the T. Thermophilus HB8 TTHA1718 protein in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Heavy metal binding protein | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5Z32
 
 | | LPS bound solution NMR structure of WS2-VR18 | | Descriptor: | VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY-LYS-ASN-LYS-SER-ARG | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
