2N7S
 
 | | Solution Structure of Leptospiral LigA4 Big Domain | | Descriptor: | Ig-like repeat domain protein 1 | | Authors: | Mei, S. | | Deposit date: | 2015-09-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of leptospiral LigA4 Big domain.
Biochem. Biophys. Res. Commun., 467, 2015
|
|
6E5X
 
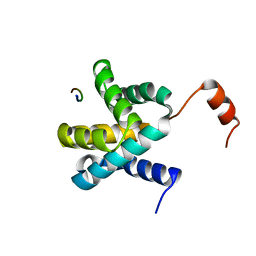 | | Crystal structure of Ebola virus VP30 C-terminus/RBBP6 peptide complex | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase RBBP6, Minor nucleoprotein VP30 | | Authors: | Liu, D, Small, G.I, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-07-23 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication.
Cell, 175, 2018
|
|
6EFG
 
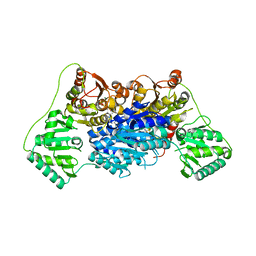 | | Pyruvate decarboxylase from Kluyveromyces lactis | | Descriptor: | MAGNESIUM ION, Pyruvate decarboxylase, THIAMINE DIPHOSPHATE | | Authors: | Kutter, S, Konig, S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structures of pyruvate decarboxylase from Kluyveromyces lactis in the absence of ligands and in the presence of the substrate surrogate pyruvamide
To be Published
|
|
6ES6
 
 | |
6ES5
 
 | |
8D6G
 
 | | Nanorana parkeri saxiphilin | | Descriptor: | PENTAETHYLENE GLYCOL, Saxiphilin | | Authors: | Zakrzewska, S, Chen, Z, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6T
 
 | | Rana catesbeiana saxiphilin mutant - Y558I:STX (co-crystal) | | Descriptor: | CACODYLATE ION, Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6O
 
 | | Nanorana parkeri saxiphilin:F-STX (soaked) | | Descriptor: | (2P)-4-({6-[({[(3aS,4R,7R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methoxy}carbonyl)amino]hexyl}carbamoyl)-2-{[4aP,9(9a)P]-6-hydroxy-3-oxo-3H-xanthen-9-yl}benzoic acid, PENTAETHYLENE GLYCOL, Saxiphilin | | Authors: | Zakrzewska, S, Chen, Z, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6P
 
 | |
8D6Q
 
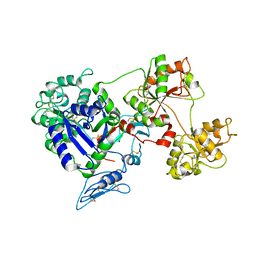 | |
8D6M
 
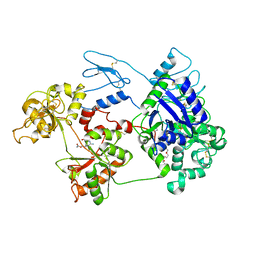 | | Nanorana parkeri saxiphilin:STX (co-crystal) | | Descriptor: | PENTAETHYLENE GLYCOL, Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Zakrzewska, S, Chen, Z, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6S
 
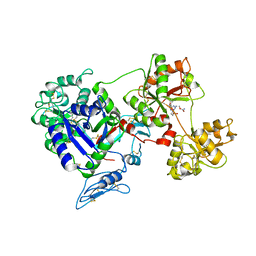 | | Rana catesbeiana saxiphilin mutant - Y558A:STX (co-crystal) | | Descriptor: | Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6U
 
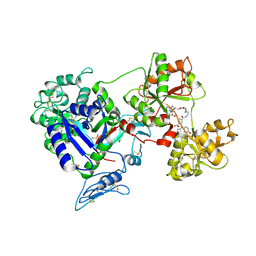 | | Rana catesbeiana saxiphilin:F-STX (soaked) | | Descriptor: | (2P)-4-({6-[({[(3aS,4R,7R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methoxy}carbonyl)amino]hexyl}carbamoyl)-2-{[4aP,9(9a)P]-6-hydroxy-3-oxo-3H-xanthen-9-yl}benzoic acid, Saxiphilin | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1BOM
 
 | | THREE-DIMENSIONAL STRUCTURE OF BOMBYXIN-II, AN INSULIN-RELATED BRAIN-SECRETORY PEPTIDE OF THE SILKMOTH BOMBYX MORI: COMPARISON WITH INSULIN AND RELAXIN | | Descriptor: | BOMBYXIN-II,BOMBYXIN A-2, BOMBYXIN-II,BOMBYXIN A-6 | | Authors: | Nagata, K, Hatanaka, H, Kohda, D, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-11-01 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of bombyxin-II an insulin-like peptide of the silkmoth Bombyx mori: structural comparison with insulin and relaxin.
J.Mol.Biol., 253, 1995
|
|
8CUY
 
 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4BV2
 
 | | CRYSTAL STRUCTURE OF SIR2 IN COMPLEX WITH THE INHIBITOR EX-527, 2'-O-ACETYL-ADP-RIBOSE AND DEACETYLATED P53-PEPTIDE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, CELLULAR TUMOR ANTIGEN P53, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8DD7
 
 | | The Cryo-EM structure of Drosophila Cryptochrome in complex with Timeless | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylated-DNA--protein-cysteine methyltransferase,Cryptochrome-1 fusion, Protein timeless,Methylated-DNA--protein-cysteine methyltransferase fusion | | Authors: | Feng, S, Lin, C, DeOliveira, C.C, Crane, B.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryptochrome-Timeless structure reveals circadian clock timing mechanisms.
Nature, 617, 2023
|
|
8D1C
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-(Trifluoromethoxy)benzoic acid | | Descriptor: | 4-(trifluoromethoxy)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2022-05-27 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Oxidations Using a Cytochrome P450 Enzyme Variant Driven with Surrogate Oxygen Donors and Light.
Chemistry, 28, 2022
|
|
8CV0
 
 | | KS-AT domains of mycobacterial Pks13 with outward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Q33
 
 | |
8CUZ
 
 | | KS-AT domains of mycobacterial Pks13 with inward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV1
 
 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6EYU
 
 | | Crystal structure of the inward H(+) pump xenorhodopsin | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Shevchenko, V, Polovinkin, V, Mager, T, Gushchin, I, Melnikov, I, Borshchevskiy, V, Popov, A, Alekseev, A, Gordeliy, V. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inward H(+) pump xenorhodopsin: Mechanism and alternative optogenetic approach.
Sci Adv, 3, 2017
|
|
6F6P
 
 | |
7Q8B
 
 | | Leishmania major actin filament in ADP-Pi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION, ... | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
