4B7J
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-20 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
8I9S
 
 | | Structure of Apo-C3aR-Go complex (Titan) | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8UKW
 
 | |
3B6U
 
 | | Crystal structure of the motor domain of human kinesin family member 3B in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF3B, MAGNESIUM ION, ... | | Authors: | Zhu, H, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Motor domain of human kinesin family member 3B in complex with ADP.
To be Published
|
|
8IA2
 
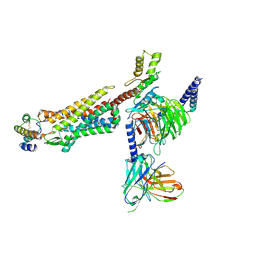 | | Structure of C5a bound human C5aR1 in complex with Go (Composite map) | | Descriptor: | Antibody fragment - ScFv16, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
4Q2Z
 
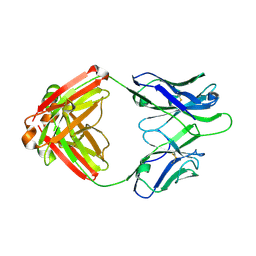 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE356, from a non-human primate | | Descriptor: | Heavy chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, Light chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody | | Authors: | Navis, M, Tran, K, Bale, S, Phad, G, Guenaga, J, Wilson, R, Soldemo, M, McKee, K, Sundling, C, Mascola, J, Li, Y, Wyatt, R.T, Hedestam, G.B.K. | | Deposit date: | 2014-04-10 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization.
Plos Pathog., 10, 2014
|
|
3PVC
 
 | |
8FJU
 
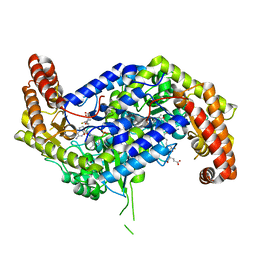 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and AGF347 inhibitor | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-{4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoyl}-L-glutamic acid, Serine hydroxymethyltransferase, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2022-12-20 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Based Design of Transport-Specific Multitargeted One-Carbon Metabolism Inhibitors in Cytosol and Mitochondria.
J.Med.Chem., 66, 2023
|
|
3M7Q
 
 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean sea anemone stichodactyla helianthus in complex with bovine pancreatic trypsin | | Descriptor: | Cationic trypsin, Kunitz-type proteinase inhibitor SHPI-1, PHOSPHATE ION | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Gil, D, Talavera, A, Gonzalez, Y, de los angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-03-17 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into serine protease inhibition by a marine invertebrate BPTI Kunitz-type inhibitor.
J.Struct.Biol., 180, 2012
|
|
2Q8E
 
 | | Specificity and Mechanism of JMJD2A, a Trimethyllysine-Specific Histone Demethylase | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Couture, J.-F, Collazo, E, Ortiz-Tello, P, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
5LUX
 
 | | Homeobox transcription factor CDX1 bound to methylated DNA | | Descriptor: | DNA (5'-D(P*GP*AP*GP*GP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*CP*CP*TP*C)-3'), ... | | Authors: | Morgunova, E, Popov, A, Taipale, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
4E6U
 
 | | Structure of LpxA from Acinetobacter baumannii at 1.4A resolution (P63 form) | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8UFH
 
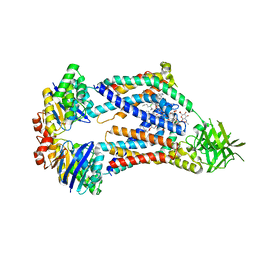 | | Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide and a macrocyclic peptide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-2-[(2~{R},4~{R},5~{R},6~{R})-5-[(2~{S},4~{R},5~{R},6~{R})-4-[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-acetamido-6-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-5-oxidanyl-oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{S})-3-dodecanoyloxydodecanoyl]oxy-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-heptanoyloxynonanoyl]amino]-3-oxidanyl-4-[(3~{R})-3-oxidanyloctanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-5-[[(3~{S})-3-[(3~{R})-3-oxidanyldodecanoyl]oxydecanoyl]amino]-3-phosphonooxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, (7S,10S,13S,17P)-10-(4-aminobutyl)-7-(3-aminopropyl)-17-(6-aminopyridin-3-yl)-20-chloro-13-[(1H-indol-3-yl)methyl]-12-methyl-6,7,9,10,12,13,15,16-octahydropyrido[2,3-b][1,5,8,11,14]benzothiatetraazacycloheptadecine-8,11,14(5H)-trione, LPS export ABC transporter permease LptG, ... | | Authors: | Pahil, K.S, Gilman, M.S.A, Baidin, V, Kruse, A.C, Kahne, D. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new antibiotic traps lipopolysaccharide in its intermembrane transporter.
Nature, 625, 2024
|
|
5OQQ
 
 | |
4AZW
 
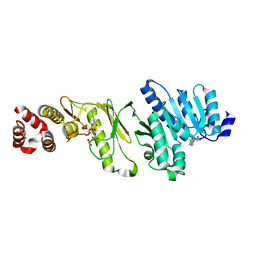 | | Crystal structure of monomeric WbdD. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
5OSP
 
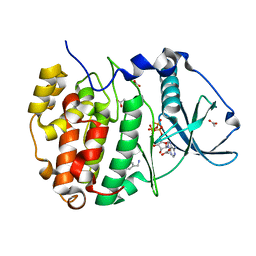 | | The crystal structure of CK2alpha in complex with an analogue of compound 1 | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-18 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
4E8C
 
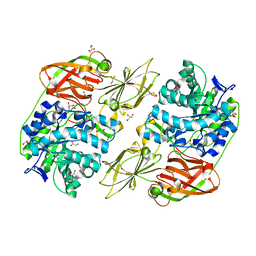 | | Crystal structure of streptococcal beta-galactosidase in complex with galactose | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35, ... | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
4MZW
 
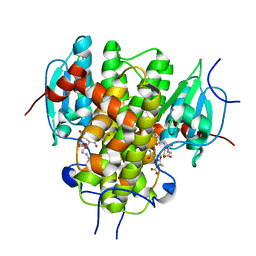 | | CRYSTAL STRUCTURE OF NU-CLASS GLUTATHIONE TRANSFERASE YGHU FROM Streptococcus sanguinis SK36, COMPLEX WITH GLUTATHIONE DISULFIDE, TARGET EFI-507286 | | Descriptor: | ACETATE ION, Glutathione S-Transferase, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Yghu (Target Efi-507286)
To be Published
|
|
3M0J
 
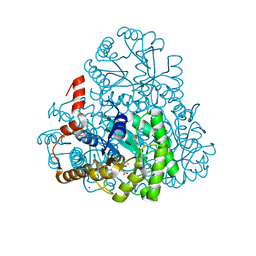 | | Structure of oxaloacetate acetylhydrolase in complex with the inhibitor 3,3-difluorooxalacetate | | Descriptor: | 2,2-difluoro-3,3-dihydroxybutanedioic acid, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Herzberg, O, Chen, C. | | Deposit date: | 2010-03-03 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus.
J.Biol.Chem., 285, 2010
|
|
1R9N
 
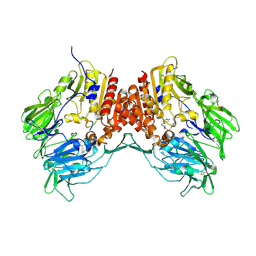 | | Crystal Structure of human dipeptidyl peptidase IV in complex with a decapeptide (tNPY) at 2.3 Ang. Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, ... | | Authors: | Aertgeerts, K, Ye, S, Tennant, M.G, Collins, B, Rogers, J, Sang, B.-C, Skene, R, Webb, D.R, Prasad, G.S. | | Deposit date: | 2003-10-30 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation.
Protein Sci., 13, 2004
|
|
3BCA
 
 | |
6XC7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.3 and CR3022 | | Descriptor: | CC12.3 heavy chain, CC12.3 light chain, CR3022 heavy chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
4N13
 
 | | Crystal Structure of PstS (BB_0215) from Borrelia burgdorferi | | Descriptor: | 1,2-ETHANEDIOL, Phosphate ABC transporter, periplasmic phosphate-binding protein, ... | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2013-10-03 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sequence, biophysical, and structural analyses of the PstS lipoprotein (BB0215) from Borrelia burgdorferi reveal a likely binding component of an ABC-type phosphate transporter.
Protein Sci., 23, 2014
|
|
5YUN
 
 | | Crystal structure of SSB complexed with myc | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Single-stranded DNA-binding protein | | Authors: | Huang, Y.H, Huang, C.Y. | | Deposit date: | 2017-11-22 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of SSB complexed with inhibitor myricetin.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6R5L
 
 | | Fragment AZ-006 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{S})-1-azanylpropan-2-yl]amino]-6-(sulfanylmethyl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
