6SI3
 
 | | p53 cancer mutant Y220S in complex with small-molecule stabilizer PK9301 | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-bromanyl-9-[2,2,2-tris(fluoranyl)ethyl]carbazol-3-yl]-~{N}-methyl-methanamine, Cellular tumor antigen p53, ... | | Authors: | Joerger, A.C, Bauer, M.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
9G10
 
 | | auxin transporter PIN8 as asymmetric dimer (outward/inward) with 4-CPA bound in the outward partly released state | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(4-chloranylphenoxy)ethanoic acid, Auxin efflux carrier component 8, ... | | Authors: | Ung, K.L, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2024-07-09 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Transport of herbicides by PIN-FORMED auxin transporters.
Biorxiv, 2024
|
|
2P69
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with PLP | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
7ACD
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound T30 (UZH1a) | | Descriptor: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-oxidanyl-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2020-09-10 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes.
Chemmedchem, 16, 2021
|
|
9G0Z
 
 | |
6SKV
 
 | |
9G0X
 
 | |
4FRS
 
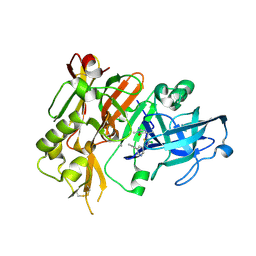 | |
7A9B
 
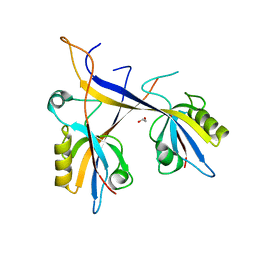 | | Crystal structure of Shank1 PDZ domain with ARAP3-derived peptide | | Descriptor: | 1,2-ETHANEDIOL, SH3 and multiple ankyrin repeat domains protein 1,Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Mariam McAuley, M, Ali, M, Ivarsson, Y, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-01 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Shank1 PDZ domain with ARAP3-derived peptide
to be published
|
|
5LPL
 
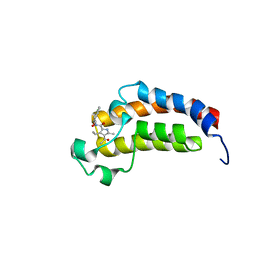 | | Crystal structure of the bromodomain of human CREBBP bound to the inhibitor XDM3c | | Descriptor: | CREB-binding protein, ~{N}-[(1~{R},2~{R})-7-chloranyl-2-oxidanyl-1,2,3,4-tetrahydronaphthalen-1-yl]-4-ethanoyl-3-ethyl-5-methyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1MN0
 
 | | Crystal structure of galactose mutarotase from lactococcus lactis complexed with D-xylose | | Descriptor: | Aldose 1-epimerase, NICKEL (II) ION, alpha-D-xylopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
6SOJ
 
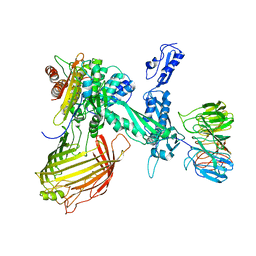 | | BamABCDE in MSP1D1 nanodisc ensemble 1-8 | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Iadanza, M.G, Ranson, N.A, Radford, S.E, Higgins, A.J, Calabrese, A.N, Schiffrin, B, White, P. | | Deposit date: | 2019-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | BamABCDE in MSP1D1 nanodisc ensemble 1-8
To Be Published
|
|
4FIX
 
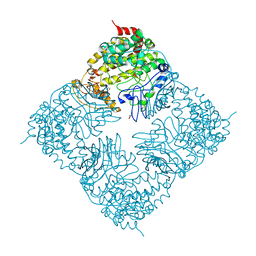 | | Crystal Structure of GlfT2 | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wheatley, R.W, Zheng, R.B, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2012-06-11 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Tetrameric Structure of the GlfT2 Galactofuranosyltransferase Reveals a Scaffold for the Assembly of Mycobacterial Arabinogalactan.
J.Biol.Chem., 287, 2012
|
|
7Y2B
 
 | |
9GGT
 
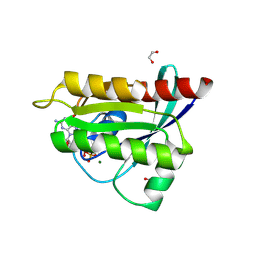 | | Human KRas4A (GDP) in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2024-08-14 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Reversible Small Molecule Multivariant Ras Inhibitors Display Tunable Affinity for the Active and Inactive Forms of Ras.
J.Med.Chem., 68, 2025
|
|
9GN8
 
 | | Crystal Structure of UFC1 E149D | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Kumar, M, Banerjee, S, Isupov, M.N, Wiener, R. | | Deposit date: | 2024-08-31 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
4FQI
 
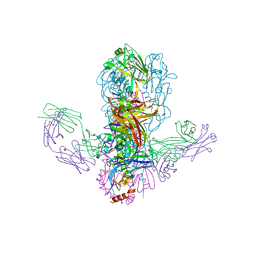 | |
5LEX
 
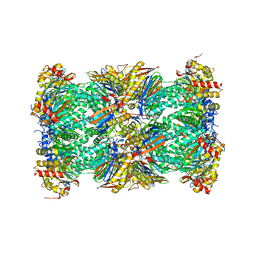 | | Native human 20S proteasome in Mg-Acetate at 2.2 Angstrom | | Descriptor: | MAGNESIUM ION, PENTAETHYLENE GLYCOL, POTASSIUM ION, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
6T1M
 
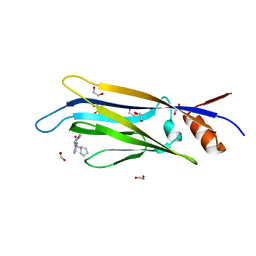 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-~{N}-[2-(piperidin-1-ylmethyl)-1~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3E7N
 
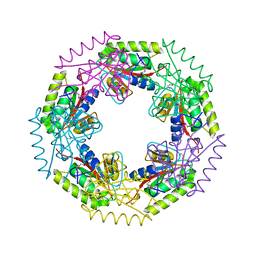 | | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2 | | Descriptor: | 1,2-ETHANEDIOL, D-ribose high-affinity transport system | | Authors: | Nocek, B, Maltseva, N, Gu, M, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-18 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2
To be Published
|
|
3E8Q
 
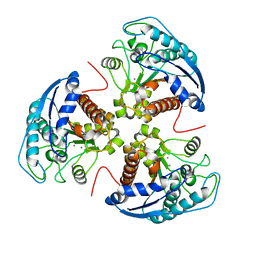 | | X-ray structure of rat arginase I-T135A: the unliganded complex | | Descriptor: | Arginase-1, MANGANESE (II) ION | | Authors: | Shishova, E.Y, Di Costanzo, L, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Probing the specificity determinants of amino acid recognition by arginase.
Biochemistry, 48, 2009
|
|
6SKS
 
 | |
8PXC
 
 | | Structure of Fap1, a domain of the accessory Sec-dependent serine-rich glycoprotein adhesin from Streptococcus oralis, solved at wavelength 3.06 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fap1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Owen, C.D, Walsh, M.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
7A9Y
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | Cellular retinoic acid-binding protein 1, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3MB7
 
 | | Human CK2 catalytic domain in complex with a difurane derivative inhibitor (AMR) | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, naphtho[2,1-b:7,8-b']difuran-2,9-dicarboxylic acid | | Authors: | Reiser, J.-B, Prudent, R, Cochet, C. | | Deposit date: | 2010-03-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Faseb J., 24, 2010
|
|
