1E54
 
 | | Anion-selective porin from Comamonas acidovorans | | Descriptor: | CALCIUM ION, OMP32, OUTER MEMBRANE PORIN PROTEIN 32, ... | | Authors: | Zeth, K, Diederichs, K, Welte, W, Engelhardt, H. | | Deposit date: | 2000-07-17 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Omp32, the Anion-Selective Porin from Comamonas Acidovorans, in Complex with a Periplasmic Peptideat 2.1 A Resolution
Structure, 8, 2000
|
|
4FST
 
 | | Crystal Structure of the CHK1 | | Descriptor: | 4-[(6,7-dimethoxy-2,4-dihydroindeno[1,2-c]pyrazol-3-yl)ethynyl]-2-methoxyphenol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FWR
 
 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-MONOPHOSPHATE, Propionate kinase | | Authors: | Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic features of Salmonella typhimurium propionate kinase (TdcD): insights from kinetic and crystallographic studies.
Biochim.Biophys.Acta, 1834, 2013
|
|
4FTJ
 
 | | Crystal Structure of the CHK1 | | Descriptor: | ISOPROPYL ALCOHOL, SULFATE ION, Serine/threonine-protein kinase Chk1, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
6SWX
 
 | | Leishmania major methionyl-tRNA synthetase in complex with an allosteric inhibitor | | Descriptor: | METHIONINE, Putative methionyl-tRNA synthetase, methyl 2-[[6-[[3,4-bis(fluoranyl)phenyl]amino]-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl]amino]ethanoate | | Authors: | Robinson, D.A, Torrie, L.S, Shepherd, S.M, De Rycker, M, Thomas, M.G, Wyatt, P.G. | | Deposit date: | 2019-09-24 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of an Allosteric Binding Site in Kinetoplastid Methionyl-tRNA Synthetase.
Acs Infect Dis., 6, 2020
|
|
4FUQ
 
 | | Crystal structure of apo MatB from Rhodopseudomonas palustris | | Descriptor: | GLYCEROL, Malonyl CoA synthetase, SULFATE ION | | Authors: | Rank, K.C, Crosby, H.A, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-06-28 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structure-Guided Expansion of the Substrate Range of Methylmalonyl Coenzyme A Synthetase (MatB) of Rhodopseudomonas palustris.
Appl.Environ.Microbiol., 78, 2012
|
|
4FWM
 
 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Propionate kinase | | Authors: | Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Mechanistic features of Salmonella typhimurium propionate kinase (TdcD): insights from kinetic and crystallographic studies.
Biochim.Biophys.Acta, 1834, 2013
|
|
5M4D
 
 | | Alpha-amino epsilon-caprolactam racemase K241A mutant in complex with D-ACL (external aldimine) | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class-III, [6-methyl-5-oxidanyl-4-[(~{E})-[(3~{R})-2-oxidanylideneazepan-3-yl]iminomethyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Frese, A, Sutton, P.W, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-10-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Snapshots of the Catalytic Cycle of the Industrial Enzyme alpha-Amino-epsilon-Caprolactam Racemase (ACLR) Observed Using X-ray Crystallography
Acs Catalysis, 7, 2017
|
|
5M4R
 
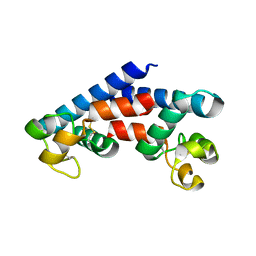 | | Structural tuning of CD81LEL (space group C2) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, SULFATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-19 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
6SXI
 
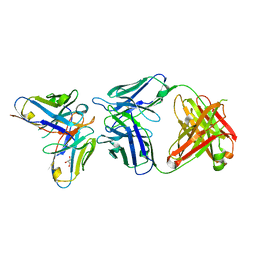 | | Antibody-anti-idiotype complex: AP33 Fab (hepatitis C virus E2 antibody) - B2.1A scFv (anti-idiotype) | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Taylor, G.L, Potter, J.A, Fadda, V, Patel, A.H, Owsianka, A.M, Cowtan, V.M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of a structural epitope mimic: an idiotypic approach to HCV vaccine design.
NPJ Vaccines, 6, 2021
|
|
2OYS
 
 | | Crystal Structure of SP1951 protein from Streptococcus pneumoniae in complex with FMN, Northeast Structural Genomics Target SpR27 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Hypothetical protein SP1951 | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Janjua, H, Satterwhite, R, Liu, J, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
4FZF
 
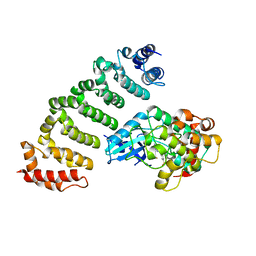 | | Crystal structure of MST4-MO25 complex with DKI | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
6SZH
 
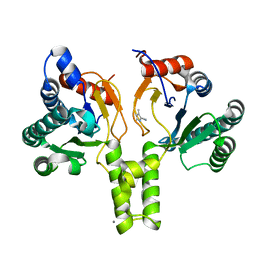 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GW197 | | Descriptor: | 3,5-dimethyl-1~{H}-pyrrole-2-carbonitrile, CALCIUM ION, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4FSQ
 
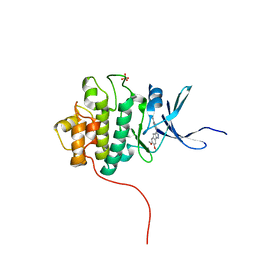 | | Crystal Structure of the CHK1 | | Descriptor: | 4'-(6,7-dimethoxyindeno[1,2-c]pyrazol-3-yl)biphenyl-4-ol, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FT0
 
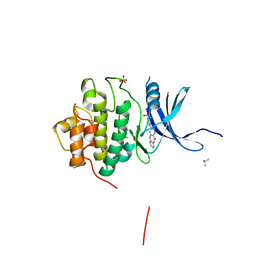 | | Crystal Structure of the CHK1 | | Descriptor: | 2-methoxy-4-(11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-3-yl)benzoic acid, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
1MDX
 
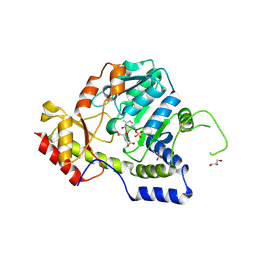 | | Crystal structure of ArnB transferase with pyridoxal 5' phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T.A, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
5M25
 
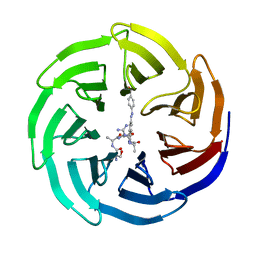 | | Modulation of MLL1 Methyltransferase Activity | | Descriptor: | (2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]propanoyl]amino]-5-carbamimidamido-~{N}-[(2~{S})-1-[[4-[(~{E})-[4-(hydroxymethyl)phenyl]diazenyl]phenyl]methylamino]-1-oxidanylidene-propan-2-yl]pentanamide, WD repeat-containing protein 5 | | Authors: | Srinivasan, V. | | Deposit date: | 2016-10-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Controlled inhibition of methyltransferases using photoswitchable peptidomimetics: towards an epigenetic regulation of leukemia.
Chem Sci, 8, 2017
|
|
6CUQ
 
 | |
6SJ4
 
 | | Amidohydrolase, AHS with substrate analog | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-hydroxyphenyl)carbonyloxybenzoic acid, Amidohydrolase, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3EEA
 
 | |
4W4O
 
 | | High-resolution crystal structure of Fc bound to its human receptor Fc-gamma-RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
5LMS
 
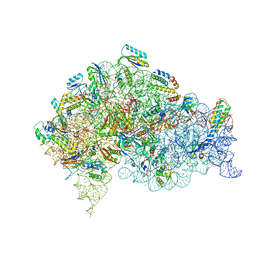 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2C) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
6N41
 
 | |
6ZT9
 
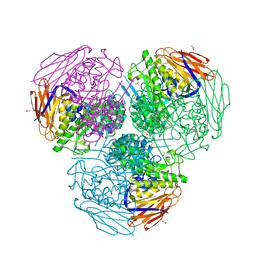 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT6
 
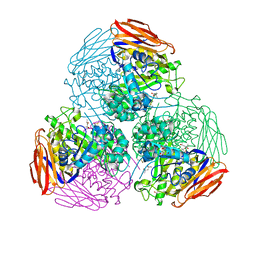 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
