7LY8
 
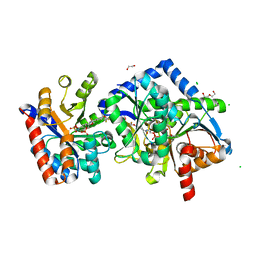 | | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with two molecules of N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the enzyme alpha-site, a single F6F molecule at the enzyme beta-site, and sodium ion at the metal coordination site at 1.55 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, CHLORIDE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-03-06 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with two molecules of N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the enzyme alpha-site, a single F6F molecule at the enzyme beta-site, and sodium ion at the metal coordination site at 1.55 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
5VIV
 
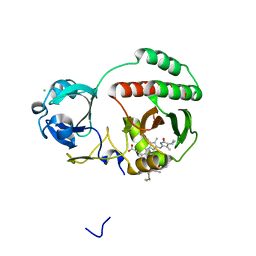 | | Crystal structure of monomeric near-infrared fluorescent protein miRFP670 | | Descriptor: | 3-[2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl] methyl]-5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]pro panoic acid, 3-[5-[[(3~{R},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | Authors: | Pletnev, S. | | Deposit date: | 2017-04-17 | | Release date: | 2017-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Designing brighter near-infrared fluorescent proteins: insights from structural and biochemical studies.
Chem Sci, 8, 2017
|
|
6VMK
 
 | |
8VWU
 
 | | Nucleosome containing 8oxoG at SHL4 | | Descriptor: | 601 I strand (damaged strand), 601 J strand (non-damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
6VP3
 
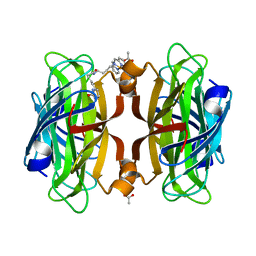 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | ACETATE ION, Streptavidin, {N-(4-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}butyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
7LWR
 
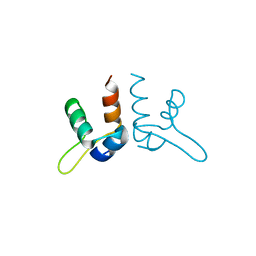 | |
8VGO
 
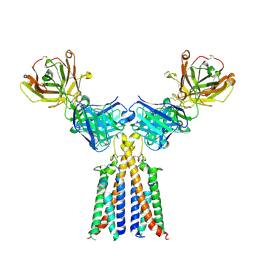 | | CryoEM structure of CD20 in complex with engineered conformationally rigid Rituximab.4DS Fab | | Descriptor: | B-lymphocyte antigen CD20, Rituximab.4DS Fab heavy chain, Rituximab.4DS Fab light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Disulfide constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
5IAT
 
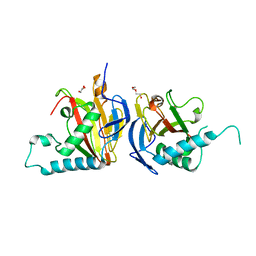 | |
8VGI
 
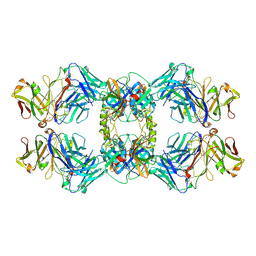 | |
8VGN
 
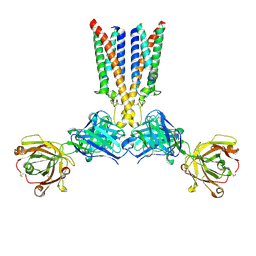 | | CryoEM structure of CD20 in complex with wild type Rituximab Fab | | Descriptor: | B-lymphocyte antigen CD20, Rituximab Fab heavy chain, Rituximab Fab light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Disulfide constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
6VRI
 
 | | Crystal Structure of the wtBlc-split Protein | | Descriptor: | Outer membrane lipoprotein Blc, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Bozhanova, N.G, Meiler, J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lipocalin Blc is a potential heme-binding protein.
Febs Lett., 595, 2021
|
|
5IKM
 
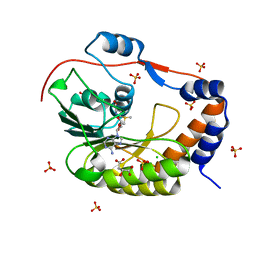 | | 1.9 Angstrom Crystal Structure of NS5 Methyl Transferase from Dengue Virus 1 in Complex with S-Adenosylmethionine and Beta-D-Fructopyranose. | | Descriptor: | CHLORIDE ION, D-MALATE, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-03 | | Release date: | 2016-03-16 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of NS5 Methyl Transferase from Dengue Virus 1 in Complex with S-Adenosylmethionine and Beta-D-Fructopyranose.
To Be Published
|
|
8VIR
 
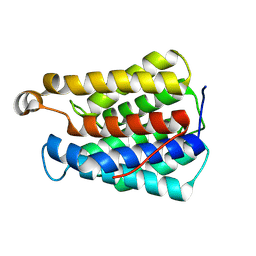 | |
5IKS
 
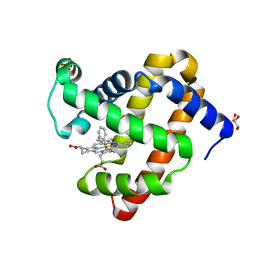 | | Wild-type sperm whale myoglobin with a Fe-phenyl moiety | | Descriptor: | GLYCEROL, Myoglobin, SULFATE ION, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2016-03-03 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Organometallic myoglobins: Formation of Fe-carbon bonds and distal pocket effects on aryl ligand conformations.
J. Inorg. Biochem., 164, 2016
|
|
8VIP
 
 | | Crystal structure of the N-terminal domain of fatty acid kinase A (FakA) from Staphylococcus aureus (Mn and AMP-PNP) | | Descriptor: | FATTY ACID KINASE A, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Bose, J.L. | | Deposit date: | 2024-01-05 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular insights into the structure and function of the Staphylococcus aureus fatty acid kinase.
J.Biol.Chem., 300, 2024
|
|
6VR6
 
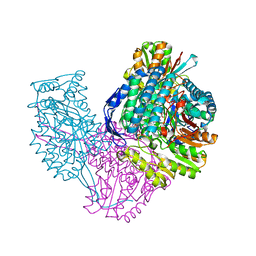 | | Structure of ALDH9A1 complexed with NAD+ in space group P1 | | Descriptor: | 4-trimethylaminobutyraldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wyatt, J.W, Tanner, J.J. | | Deposit date: | 2020-02-06 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch.Biochem.Biophys., 691, 2020
|
|
5VO2
 
 | |
5IKU
 
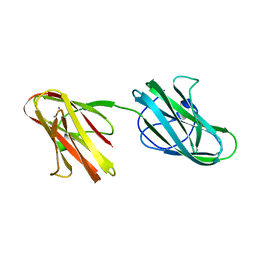 | | Crystal structure of the Hathewaya histolytica ColG tandem collagen-binding domain s3as3b in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, Collagenase | | Authors: | Janowska, K, Bauer, R, Roeser, R, Sakon, J, Matsushita, O. | | Deposit date: | 2016-03-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca2+-induced orientation of tandem collagen binding domains from clostridial collagenase ColG permits two opposing functions of collagen fibril formation and retardation.
Febs J., 285, 2018
|
|
8VJE
 
 | | Human R14A Pin1 covalently bound to inhibitor 158F10 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Inhibitor 158F10, ... | | Authors: | Rodriguez, I, Blaha, G. | | Deposit date: | 2024-01-06 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeted degradation of Pin1 by protein-destabilizing compounds.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VIQ
 
 | | Crystal structure of the N-terminal domain of fatty acid kinase A (FakA) from Staphylococcus aureus (Mg and AMP-PNP) | | Descriptor: | FATTY ACID KINASE A, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Bose, J.L. | | Deposit date: | 2024-01-05 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular insights into the structure and function of the Staphylococcus aureus fatty acid kinase.
J.Biol.Chem., 300, 2024
|
|
8VWJ
 
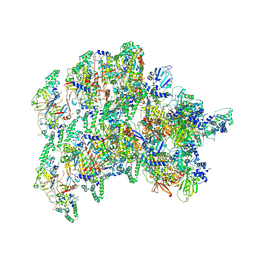 | | The base complex of the AcMNPV baculovirus nucleocapsid (Class 2, localised reconstruction) | | Descriptor: | 38K (AC98), Capsid-associated protein VP80, Major capsid protein, ... | | Authors: | Johnstone, B.A, Koszalka, P, Ha, J, Venugopal, H, Coulibaly, F. | | Deposit date: | 2024-02-01 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.78 Å) | | Cite: | The nucleocapsid architecture and structural atlas of the prototype baculovirus define the hallmarks of a new viral realm.
Sci Adv, 10, 2024
|
|
5VKK
 
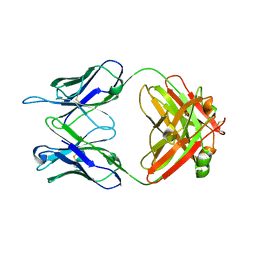 | |
5IBY
 
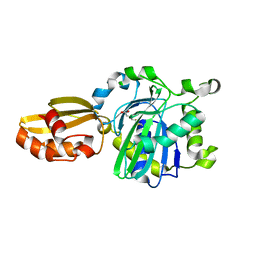 | |
8VJF
 
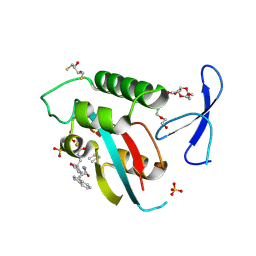 | | Human R14A Pin1 covalently bound to inhibitor 158D9 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Rodriguez, I, Blaha, G. | | Deposit date: | 2024-01-06 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeted degradation of Pin1 by protein-destabilizing compounds.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VWH
 
 | | Structure of the baculovirus major nucleocapsid protein VP39 (localised reconstruction) | | Descriptor: | Major capsid protein, ZINC ION | | Authors: | Johnstone, B.A, Hardy, J.M, Ha, J.H, Venugopal, H, Coulibaly, F. | | Deposit date: | 2024-02-01 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The nucleocapsid architecture and structural atlas of the prototype baculovirus define the hallmarks of a new viral realm.
Sci Adv, 10, 2024
|
|
