7O4P
 
 | | Cystal structure of Zymogen Granule Protein 16 (ZG16) | | Descriptor: | CHLORIDE ION, GLYCEROL, Zymogen granule membrane protein 16 | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2021-04-07 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Conformational switches and redox properties of the colon cancer-associated human lectin ZG16.
Febs J., 288, 2021
|
|
7O3I
 
 | | Cystal structure of Zymogen Granule Protein 16 (ZG16) | | Descriptor: | CHLORIDE ION, GLYCEROL, Zymogen granule membrane protein 16 | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational switches and redox properties of the colon cancer-associated human lectin ZG16.
Febs J., 288, 2021
|
|
7O88
 
 | |
5OM7
 
 | |
6HGH
 
 | |
6HGE
 
 | |
6HGI
 
 | |
6HGD
 
 | |
6HGL
 
 | |
1M21
 
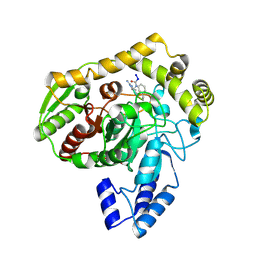 | | Crystal structure analysis of the peptide amidase PAM in complex with the competitive inhibitor chymostatin | | Descriptor: | CHYMOSTATIN, Peptide Amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
1P0E
 
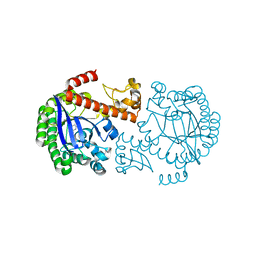 | | CRYSTAL STRUCTURE OF ZYMOMONAS MOBILIS tRNA-GUANINE TRANSGLYCOSYLASE (TGT) COCRYSTALLISED WITH PREQ1 AT PH 5.5 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase
and their implications for substrate selectivity, reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
5NEY
 
 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, CYS-TRD-TRP-LYD-LYS-LYD-LYS-LYD-TRP-TRD-CYS-ALA, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
5NF0
 
 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, CYD-TRP-TRD-LYS-LYD-LYS-LYD-LYS-TRD-TRP-CYD-GLY, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
5NES
 
 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 1,3-dimethylbenzene, 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
6Q40
 
 | | A secreted LysM effector of the wheat pathogen Zymoseptoria tritici protects the fungal hyphae against chitinase hydrolysis through ligand-dependent polymerisation of LysM homodimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, LysM domain-containing protein | | Authors: | Mesters, J.R, Saleem-Batcha, R, Sanchez-Vallet, A, Thomma, B.P.H.J. | | Deposit date: | 2018-12-05 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization.
Plos Pathog., 16, 2020
|
|
1P0D
 
 | | CRYSTAL STRUCTURE OF ZYMOMONAS MOBILIS tRNA-GUANINE TRANSGLYCOSYLASE (TGT) CRYSTALLISED AT PH 5.5 | | Descriptor: | Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme
tRNA-guanine transglycosylase and their implications for substrate selectivity,
reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
4BIM
 
 | |
8TXY
 
 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3ZJ4
 
 | |
3ZJ5
 
 | | NEUROSPORA CRASSA CATALASE-3 EXPRESSED IN E. COLI, ORTHORHOMBIC FORM. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, CATALASE-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E. | | Deposit date: | 2013-01-17 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conformational Stability and Crystal Packing: Polymorphism in Neurospora Crassa Cat-3
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1P0B
 
 | | Crystal Structure Of tRNA-Guanine Transglycosylase (TGT) From Zymomonas mobilis Complexed With Archaeosine Precursor, Preq0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase
and their implications for substrate selectivity, reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
4A3U
 
 | |
4IEF
 
 | | Complex of Porphyromonas gingivalis RgpB pro- and mature domains | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BARIUM ION, CALCIUM ION, ... | | Authors: | de Diego, I, Veillard, F.T, Guevara, T, Potempa, B, Sztukowska, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Porphyromonas gingivalis Virulence Factor Gingipain RgpB Shows a Unique Zymogenic Mechanism for Cysteine Peptidases.
J.Biol.Chem., 288, 2013
|
|
4G3G
 
 | |
5UG0
 
 | | Human antibody H2897 in complex with influenza hemagglutinin H1 Solomon Islands/03/2006 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2897 heavy chain, ... | | Authors: | Raymond, D.D, Caradonna, T, Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2017-01-06 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
