3TSZ
 
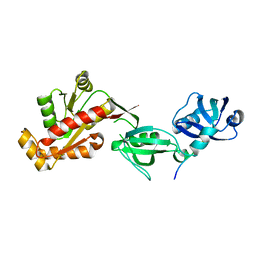 | |
1L1K
 
 | |
1G5K
 
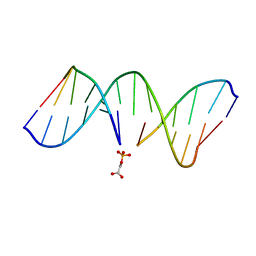 | | NMR Structrure of d(CCAAAGXACTGGG), X is a 3'-phosphoglycolate, 5'-phosphate gapped lesion, 10 structures | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*CP*AP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3', ... | | Authors: | Junker, H.-D, Hoehn, S.T, Bunt, R.C, Marathius, V, Chen, J, Turner, C.J, Stubbe, J. | | Deposit date: | 2002-10-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Characterization and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Nucleic Acids Res., 30, 2002
|
|
1FU1
 
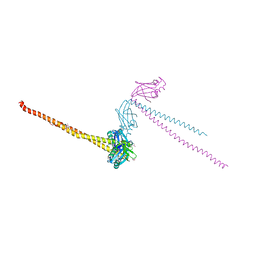 | | CRYSTAL STRUCTURE OF HUMAN XRCC4 | | Descriptor: | ACETIC ACID, DNA REPAIR PROTEIN XRCC4 | | Authors: | Junop, M, Modesti, M, Guarne, A, Gellert, M, Yang, W. | | Deposit date: | 2000-09-13 | | Release date: | 2000-12-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Xrcc4 DNA repair protein and implications for end joining.
EMBO J., 19, 2000
|
|
1GJ1
 
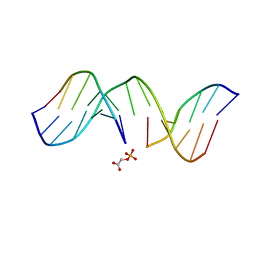 | | NMR structure of d(CCAAAGXACTGGG), X is a 3'phosphoglycolate, 5'phosphate gapped lesion | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*CP*AP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3', ... | | Authors: | Junker, H.-D, Hoehn, S.T, Bunt, R.C, Marathius, V, Chen, J, Turner, C.J, Stubbe, J. | | Deposit date: | 2002-10-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Characterization and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Nucleic Acids Res., 30, 2002
|
|
1LU8
 
 | |
2LRL
 
 | | Solution Structures of the IIA(Chitobiose)-HPr complex of the N,N'-Diacetylchitobiose Branch of the Escherichia coli Phosphotransferase System | | Descriptor: | N,N'-diacetylchitobiose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphocarrier protein HPr | | Authors: | Jung, Y, Cai, M, Clore, M. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the IIAChitobiose-HPr Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia coli Phosphotransferase System.
J.Biol.Chem., 287, 2012
|
|
1RP7
 
 | | E. COLI PYRUVATE DEHYDROGENASE INHIBITOR COMPLEX | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Arjunan, P, Chandrasekhar, K, Furey, W. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Determinants of Enzyme Binding Affinity: The E1 Component of Pyruvate Dehydrogenase from Escherichia coli in Complex with the Inhibitor Thiamin Thiazolone Diphosphate.
Biochemistry, 43, 2004
|
|
1HR2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT P4-P6 DOMAIN (DELC209) OF TETRAHYMENA THEMOPHILA GROUP I INTRON. | | Descriptor: | MAGNESIUM ION, P4-P6 DELC209 MUTANT RNA RIBOZYME DOMAIN | | Authors: | Juneau, K, Podell, E.R, Harrington, D.J, Cech, T.R. | | Deposit date: | 2000-12-20 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions.
Structure, 9, 2001
|
|
2NT2
 
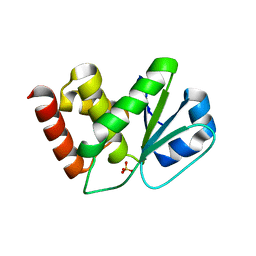 | | Crystal Structure of Slingshot phosphatase 2 | | Descriptor: | Protein phosphatase Slingshot homolog 2, SULFATE ION | | Authors: | Jung, S.K, Jeong, D.G, Yoon, T.S, Kim, J.H, Ryu, S.E, Kim, S.J. | | Deposit date: | 2006-11-06 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human slingshot phosphatase 2.
Proteins, 68, 2007
|
|
2ORY
 
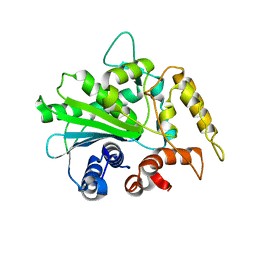 | | Crystal structure of M37 lipase | | Descriptor: | Lipase | | Authors: | Jung, S.K, Jeong, D.G, Kim, S.J. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum.
Proteins, 71, 2008
|
|
6W5X
 
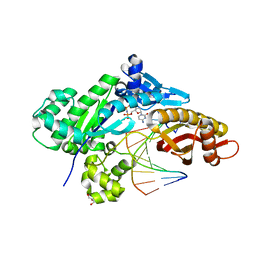 | | Crystal structure of human polymerase eta complexed with N7-benzylguanine and dCTP* | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*AP*TP*(DRG)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of human polymerase eta complexed with N7-benzylguanine and dCTP*
To Be Published
|
|
384D
 
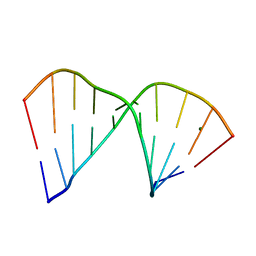 | |
329D
 
 | |
382D
 
 | |
2KFB
 
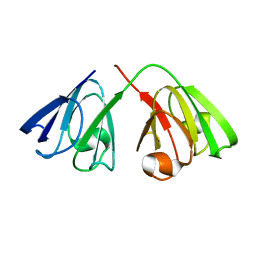 | | The structure of the cataract causing P23T mutant of human gamma-D crystallin | | Descriptor: | Gamma-crystallin D | | Authors: | Jung, J, Byeon, I.L, Wang, Y, King, J, Gronenborn, A.M. | | Deposit date: | 2009-02-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the cataract-causing P23T mutant of human gammaD-crystallin exhibits distinctive local conformational and dynamic changes.
Biochemistry, 48, 2009
|
|
383D
 
 | |
2KSG
 
 | |
2IYG
 
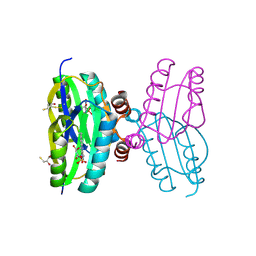 | | Dark state structure of the BLUF domain of the rhodobacterial protein AppA | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, APPA, ... | | Authors: | Jung, A, Reinstein, J, Domratcheva, T, Shoeman, R.-L, Schlichting, I. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Appa Bluf Domain Photoreceptor Provide Insights Into Blue Light- Mediated Signal Transduction.
J.Mol.Biol., 362, 2006
|
|
6WK6
 
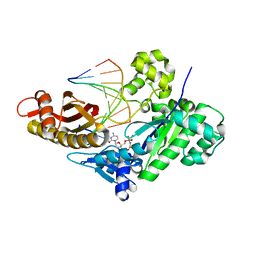 | | Crystal structure of human polymerase eta complexed with Xanthine containing DNA | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*AP*TP*(3ZO)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-04-15 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the bypass of the major deaminated purines by translesion synthesis DNA polymerase.
Biochem.J., 477, 2020
|
|
2IYI
 
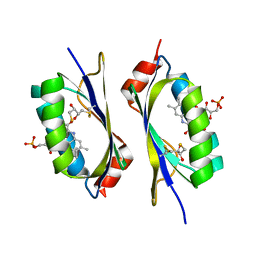 | | structure of a light-induced intermediate of the BLUF domain of the rhodobacterial protein AppA | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, APPA, ... | | Authors: | Jung, A, Reinstein, J, Domratcheva, T, Shoeman, R.-L, Schlichting, I. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-06 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction.
J. Mol. Biol., 362, 2006
|
|
2LLR
 
 | |
2LN8
 
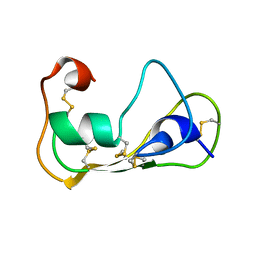 | | The solution structure of theromacin | | Descriptor: | Theromacin | | Authors: | Jung, S, Soennichsen, F.D, Hung, C.-W, Tholey, A, Boidin-Wichlacz, C, Hausgen, W, Gelhaus, C, Desel, C, Podschun, R, Watzig, V, Tasiemski, A, Leippe, M, Groetzinger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Macin family of antimicrobial proteins combines antimicrobial and nerve repair activities.
J.Biol.Chem., 287, 2012
|
|
2K35
 
 | | Hydramacin-1: Structure and antibacterial activity of a peptide from the basal metazoan Hydra | | Descriptor: | hydramacin-1 | | Authors: | Jung, S, Dingley, A.J, Stanisak, M, Gelhaus, C, Bosch, T, Podschun, R, Leippe, M, Gr tzinger, J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Hydramacin-1, structure and antibacterial activity of a protein from the Basal metazoan hydra.
J.Biol.Chem., 284, 2009
|
|
1F3R
 
 | | COMPLEX BETWEEN FV ANTIBODY FRAGMENT AND AN ANALOGUE OF THE MAIN IMMUNOGENIC REGION OF THE ACETYLCHOLINE RECEPTOR | | Descriptor: | ACETYLCHOLINE RECEPTOR ALPHA, FV ANTIBODY FRAGMENT | | Authors: | Kleinjung, J, Petit, M.-C, Orlewski, P, Mamalaki, A, Tzartos, S.-J, Tsikaris, V, Sakarellos-Daitsiotis, M, Sakarellos, C, Marraud, M, Cung, M.-T. | | Deposit date: | 2000-06-06 | | Release date: | 2000-06-15 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The third-dimensional structure of the complex between an Fv antibody fragment and an analogue of the main immunogenic region of the acetylcholine receptor: a combined two-dimensional NMR, homology, and molecular modeling approach.
Biopolymers, 53, 2000
|
|
