7OVF
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 8 (JMV-7207) | | Descriptor: | 4-[2-[(4-fluorophenyl)methylsulfanyl]ethyl]-3-phenyl-1H-1,2,4-triazole-5-thione, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Marcoccia, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
8FAJ
 
 | | OXA-48-NA-1-157 inhibitor complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Toth, M, Vakulenko, S.B. | | Deposit date: | 2022-11-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Exhibits Potent Activity against Klebsiella spp. Isolates Producing OXA-48-Type Carbapenemases.
Acs Infect Dis., 9, 2023
|
|
1FP0
 
 | | SOLUTION STRUCTURE OF THE PHD DOMAIN FROM THE KAP-1 COREPRESSOR | | Descriptor: | KAP-1 COREPRESSOR, ZINC ION | | Authors: | Capili, A.D, Schultz, D.C, Rauscher III, F.J, Borden, K.L.B. | | Deposit date: | 2000-08-29 | | Release date: | 2001-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains.
EMBO J., 20, 2001
|
|
1FRP
 
 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-2,6-BISPHOSPHATE, AMP AND ZN2+ AT 2.0 ANGSTROMS RESOLUTION. ASPECTS OF SYNERGISM BETWEEN INHIBITORS | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Xue, Y, Huang, S, Liang, J.-Y, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-08-26 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 2,6-bisphosphate, AMP, and Zn2+ at 2.0-A resolution: aspects of synergism between inhibitors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1FBP
 
 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE 6-PHOSPHATE, AMP, AND MAGNESIUM | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Ke, H, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1990-05-31 | | Release date: | 1992-04-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 6-phosphate, AMP, and magnesium.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1FPB
 
 | | CRYSTAL STRUCTURE OF THE NEUTRAL FORM OF FRUCTOSE 1,6-BISPHOSPHATASE COMPLEXED WITH REGULATORY INHIBITOR FRUCTOSE 2,6-BISPHOSPHATE AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Liang, J.Y, Huang, S, Zhang, Y, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the neutral form of fructose 1,6-bisphosphatase complexed with regulatory inhibitor fructose 2,6-bisphosphate at 2.6-A resolution.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1FTA
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE, 1-PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH THE ALLOSTERIC INHIBITOR AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Lipscomb, W.N. | | Deposit date: | 1993-09-27 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric site of human liver fructose-1,6-bisphosphatase. Analysis of six AMP site mutants based on the crystal structure.
J.Biol.Chem., 269, 1994
|
|
8GPZ
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with C239-0012 | | Descriptor: | 3-methyl-6-(4-methylpiperidin-1-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2022-08-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
8ZKR
 
 | |
1FUA
 
 | | L-FUCULOSE 1-PHOSPHATE ALDOLASE CRYSTAL FORM T | | Descriptor: | BETA-MERCAPTOETHANOL, L-FUCULOSE-1-PHOSPHATE ALDOLASE, SULFATE ION, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Refined high-resolution structure of the metal-ion dependent L-fuculose-1-phosphate aldolase (class II) from Escherichia coli.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1G9M
 
 | | HIV-1 HXBC2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Kwong, P.D, Wyatt, R, Majeed, S, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 2000-11-24 | | Release date: | 2000-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates.
Structure Fold.Des., 8, 2000
|
|
7PHL
 
 | | Human voltage-gated potassium channel Kv3.1 (with EDTA) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Qian, P, Castro-Hartmann, P, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Fernandez-Cid, A, Pike, A.C.W, Marsden, B, MacLean, E.M, Sader, K, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7PHI
 
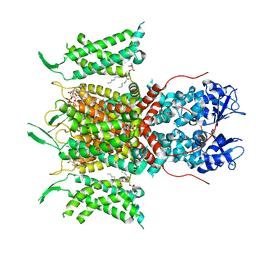 | | Human voltage-gated potassium channel Kv3.1 (with Zn) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Qian, P, Castro-Hartmann, P, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Fernandez-Cid, A, Marsden, B, MacLean, E.M, Pike, A.C.W, Sader, K, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7PHK
 
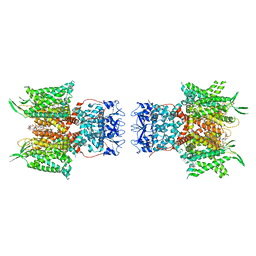 | | Human voltage-gated potassium channel Kv3.1 in dimeric state (with Zn) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Qian, P, Castro-Hartmann, P, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Marsden, B, MacLean, E.M, Fernandez-Cid, A, Pike, A.C.W, Sader, K, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7PP0
 
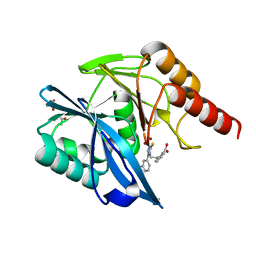 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in complex with compound 28 (JMV-7038) | | Descriptor: | 2-[2-[3-[3-(2-morpholin-4-ylethoxy)phenyl]-5-sulfanylidene-1H-1,2,4-triazol-4-yl]ethyl]benzoic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Corsica, G, Chelini, G, De Luca, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1,2,4-Triazole-3-Thione Analogues with a 2-Ethylbenzoic Acid at Position 4 as VIM-type Metallo-beta-Lactamase Inhibitors.
Chemmedchem, 17, 2022
|
|
7O2F
 
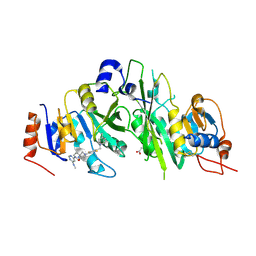 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 22 (UZH2) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]-2,5-bis(fluoranyl)phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8EK9
 
 | | Crystal structure of the class A carbapenemase CRH-1 in complex with avibactam at 1.4 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Brunetti, F, Ghiglione, B, Guardabassi, L, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of CRH-1, a Carbapenemase from Chromobacterium haemolyticum Related to KPC beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
7O2E
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 21 (ADO_AD_089) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]-3-fluoranyl-phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O09
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 7 (ADO_AC_074) | | Descriptor: | 6-[4-[6-[(4,4-dimethylpiperidin-1-yl)methyl]pyridin-3-yl]-1-oxa-4,9-diazaspiro[5.5]undecan-9-yl]-N-(phenylmethyl)pyrimidin-4-amine, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O08
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 5 (ADO_AB_075) | | Descriptor: | 4-[[[6-[(4,4-dimethylpiperidin-1-yl)methyl]pyridin-3-yl]amino]methyl]-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-4-ol, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O0L
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 8 (ADO_AC_093) | | Descriptor: | 4-[6-[(4,4-dimethylpiperidin-1-yl)methyl]pyridin-3-yl]-9-[6-[(phenylmethyl)amino]pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O29
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 20 (ADO_AD_044) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-fluoranyl-phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8EHH
 
 | | Crystal structure of the class A extended-spectrum beta-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Ghiglione, B, Bonomo, R.A, Rodriguez, M.M, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Crystal structure of the class A extended-spectrum beta-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution.
Antimicrob.Agents Chemother., 68, 2024
|
|
6VOS
 
 | | Crystal structure of macaque anti-HIV-1 antibody RM20J | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
7OSK
 
 | | Ignisphaera aggregans GH53 catalytic domain | | Descriptor: | Arabinogalactan endo-1,4-beta-galactosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fredslund, F, Lo Leggio, L, Poulsen, J.C, Rasmussen, K.K, Muderspach, S, Krogh, K.B.R.M, Jensen, K. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineering the substrate binding site of the hyperthermostable archaeal endo-beta-1,4-galactanase from Ignisphaera aggregans.
Biotechnol Biofuels, 14, 2021
|
|
