5I8U
 
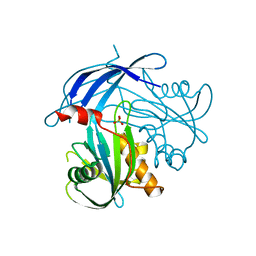 | | Crystal Structure of the RV1700 (MT ADPRASE) E142Q mutant | | Descriptor: | ADP-ribose pyrophosphatase, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Thirawatananond, P, Kang, L.-W, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis.
J. Bioenerg. Biomembr., 48, 2016
|
|
5VDP
 
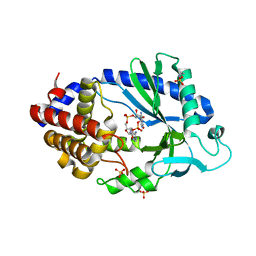 | |
6VTH
 
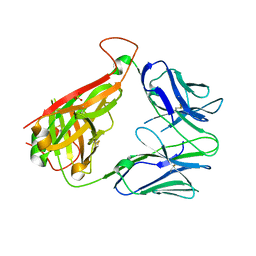 | | p53-specific T cell receptor | | Descriptor: | SULFATE ION, T-cell Receptor 12-6, Alfa chain, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
8VK9
 
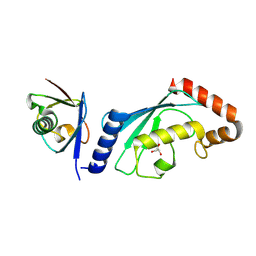 | | Structure of UbV.d2.3 in complex with Ube2d2-S22R | | Descriptor: | GLYCEROL, Ubiquitin variant D2.3, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Middleton, A.J. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-25 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biophysical characterisation of ubiquitin variants that inhibit the ubiquitin conjugating enzyme Ube2d2.
Febs J., 291, 2024
|
|
6VIW
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
8VQK
 
 | | YcjN from Escherichia coli | | Descriptor: | CADMIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Trevino, M.A, Fernandez, D, Sharaf, N.G. | | Deposit date: | 2024-01-18 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression, purification, and characterization of diacylated Lipo-YcjN from Escherichia coli.
J.Biol.Chem., 300, 2024
|
|
6VJ2
 
 | | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis | | Descriptor: | Foldase | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis
To Be Published
|
|
8VU3
 
 | | Cryo-EM structure of cyanobacterial PSI with bound platinum nanoparticles | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Gisriel, C.J, Malavath, T, Brudvig, G.W, Utschig, L.M. | | Deposit date: | 2024-01-28 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Structure of a biohybrid photosystem I-platinum nanoparticle solar fuel catalyst.
Nat Commun, 15, 2024
|
|
5I9O
 
 | |
8VX4
 
 | | Human OGG1 bound to a 35-bp DNA with an 8-oxoG in the middle | | Descriptor: | DNA (35-MER), N-glycosylase/DNA lyase | | Authors: | You, Q, Li, H. | | Deposit date: | 2024-02-03 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations.
Commun Biol, 7, 2024
|
|
7LKL
 
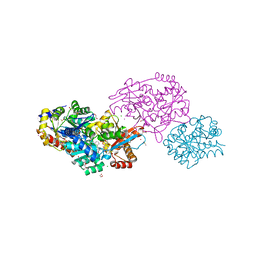 | | The internal aldimine form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site and the product L-tryptophan at the enzyme beta-site at 1.05 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-02-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site and the product L-tryptophan at the enzyme beta-site at 1.05 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
8VOZ
 
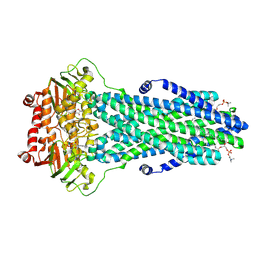 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with MgADP and substrate | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxypropyl hexadecanoate, ABC-type bacteriocin transporter, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
5VN6
 
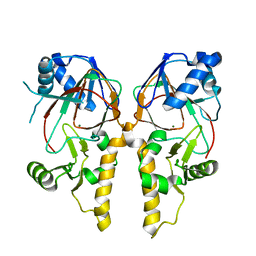 | |
8VJA
 
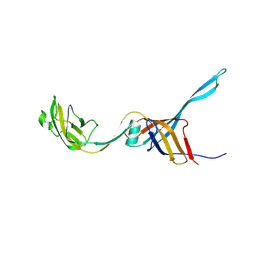 | |
7LI5
 
 | | Crystal Structure Analysis of human TEAD1 | | Descriptor: | 1-[(3R,4R)-3-[4-(pyridin-3-yl)-1H-1,2,3-triazol-1-yl]-4-{[4-(trifluoromethyl)phenyl]methoxy}pyrrolidin-1-yl]prop-2-en-1-one, SULFATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure Analysis of human TEAD1
To Be Published
|
|
5IAW
 
 | | Novel natural FXR modulator with a unique binding mode | | Descriptor: | (1S,2R,4S)-1,7,7-trimethylbicyclo[2.2.1]heptan-2-yl 4-hydroxybenzoate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Novel Class of Natural FXR Modulators with a Unique Mode of Selective Co-regulator Assembly
Chembiochem, 18, 2017
|
|
8VDO
 
 | |
8VDP
 
 | |
5IB7
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet, near-cognate tRNALys with U-G mismatch in the A-site and antibiotic paromomycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
8VJM
 
 | | SpoIVFB(E44Q variant):pro-sigmaK complex | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Lauryl Maltose Neopentyl Glycol, RNA polymerase sigma-K factor, ... | | Authors: | Orlando, M.A, Pouillon, H.J.T, Mandal, S, Kroos, L, Orlando, B.J. | | Deposit date: | 2024-01-07 | | Release date: | 2024-10-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Substrate engagement by the intramembrane metalloprotease SpoIVFB.
Nat Commun, 15, 2024
|
|
7LK0
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) | | Descriptor: | (1R,3S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Shen, S, Liu, D, Silverman, R. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
8VJI
 
 | | Cryo-EM of capsid of bacteriophage Chi | | Descriptor: | Decorator protein D, Major capsid protein | | Authors: | Sonani, R.R, Esteves, N.C, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2024-01-06 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of flagellotropic bacteriophage Chi.
Structure, 32, 2024
|
|
8VPV
 
 | | Class III PreQ1 riboswitch mutant delta84 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, RNA (101-MER) | | Authors: | Srivastava, Y, Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2024-01-17 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Two riboswitch classes that share a common ligand-binding fold show major differences in the ability to accommodate mutations.
Nucleic Acids Res., 52, 2024
|
|
5VHB
 
 | |
6VN6
 
 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 14 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION, [(2R)-5-chloro-7-{2-[(2S)-1-chloro-2,3-dihydroxypropan-2-yl]thieno[3,2-b]pyridin-7-yl}-2,3-dihydro-1-benzofuran-2-yl](piperazin-1-yl)methanone | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
