1LP1
 
 | | Protein Z in complex with an in vitro selected affibody | | Descriptor: | Affibody binding protein Z, Immunoglobulin G binding protein A, MAGNESIUM ION, ... | | Authors: | Hogbom, M, Eklund, M, Nygren, P.A, Nordlund, P. | | Deposit date: | 2002-05-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition by an in vitro evolved affibody.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1GWT
 
 | |
1QV7
 
 | | HORSE LIVER ALCOHOL DEHYDROGENASE HIS51GLN/LYS228ARG MUTANT COMPLEXED WITH NAD+ AND 2,3-DIFLUOROBENZYL ALCOHOL | | Descriptor: | 2,3-DIFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lebrun, L.A, Park, D.-H, Ramaswamy, S, Plapp, B.V. | | Deposit date: | 2003-08-27 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase.
Biochemistry, 43, 2004
|
|
1HJU
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HXY
 
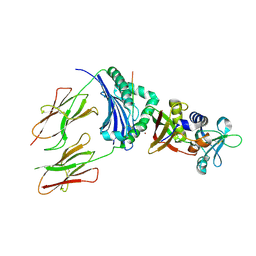 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCAL ENTEROTOXIN H IN COMPLEX WITH HUMAN MHC CLASS II | | Descriptor: | ENTEROTOXIN H, HEMAGGLUTININ, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Petersson, K, Hakansson, M, Nilsson, H, Forsberg, G, Svensson, L.A, Liljas, A, Walse, B. | | Deposit date: | 2001-01-17 | | Release date: | 2001-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Superantigen Bound to MHC Class II Displays Zinc and Peptide Dependence
Embo J., 20, 2001
|
|
1QKC
 
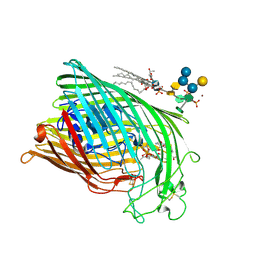 | | ESCHERICHIA COLI FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) IN COMPLEX DELTA TWO-ALBOMYCIN | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DELTA-2-ALBOMYCIN A1, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Braun, V, Fiedler, H.-P, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-07-18 | | Release date: | 2000-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA.
Protein Sci., 9, 2000
|
|
1NNP
 
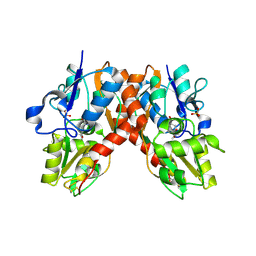 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.9 A resolution. Crystallization without zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, Glutamate receptor 2, SULFATE ION | | Authors: | Lunn, M.L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
1HXJ
 
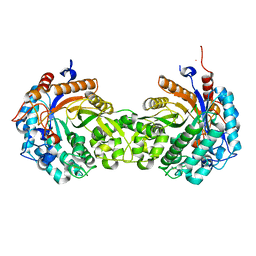 | |
1HJQ
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1JQP
 
 | | dipeptidyl peptidase I (cathepsin C), a tetrameric cysteine protease of the papain family | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Olsen, J.G, Kadziola, A, Lauritzen, C, Pedersen, J, Larsen, S, Dahl, S.W. | | Deposit date: | 2001-08-08 | | Release date: | 2002-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tetrameric dipeptidyl peptidase I directs substrate specificity by use of the residual pro-part domain
FEBS LETT., 506, 2001
|
|
3HV6
 
 | | Human p38 MAP Kinase in Complex with RL39 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[4-(2-morpholin-4-ylethoxy)phenyl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
1NNK
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.85 A resolution. Crystallization with zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Lunn, M.-L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
1JF2
 
 | | Crystal Structure of W92F obelin mutant from Obelia longissima at 1.72 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, obelin | | Authors: | Liu, Z.-J, Vysotski, E.S, Deng, L, Markova, S.V, Lee, J, Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Violet bioluminescence and fast kinetics from W92F obelin: structure-based proposals for the bioluminescence triggering and the identification of the emitting species.
Biochemistry, 42, 2003
|
|
1JRB
 
 | | The P56A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, OROTIC ACID, dihydroorotate dehydrogenase A | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-13 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
8CQ2
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, C16S, C20S, C870S, T1279C mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Multistate kinetics of the syringe-like injection mechanism of Tc toxins.
Sci Adv, 11, 2025
|
|
8CPZ
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K1179W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Multistate kinetics of the syringe-like injection mechanism of Tc toxins.
Sci Adv, 11, 2025
|
|
8CQ0
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K567W K2008W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Multistate kinetics of the syringe-like injection mechanism of Tc toxins.
Sci Adv, 11, 2025
|
|
1RXG
 
 | | DEACETOXYCEPHALOSPORIN C SYNTHASE COMPLEXED WITH FE(II) AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION, ... | | Authors: | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Shofield, C.J, Hajdu, J, Andersson, I. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
2IZM
 
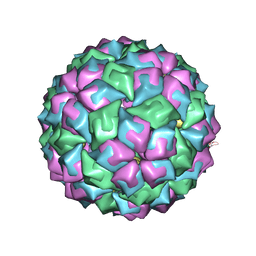 | | MS2-RNA HAIRPIN (C-10) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*CP*GP*GP*AP*UP *CP*AP*CP*CP*CP*AP*UP*GP*U)-3', Capsid protein | | Authors: | Helgstrand, C, Grahn, E, Moss, T, Stonehouse, N.J, Tars, K, Stockley, P.G, Liljas, L. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Investigating the Structural Basis of Purine Specificity in the Structures of MS2 Coat Protein RNA Translational Operator Complexes
Nucleic Acids Res., 30, 2002
|
|
2J74
 
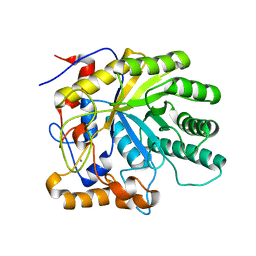 | | Structure of Beta-1,4-Galactanase | | Descriptor: | CALCIUM ION, YVFO, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Le Nours, J, De Maria, L, Welner, D, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-10-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigating the Binding of Beta-1,4-Galactan to Bacillus Licheniformis Beta-1,4-Galactanase by Crystallography and Computational Modeling.
Proteins, 75, 2009
|
|
1SJW
 
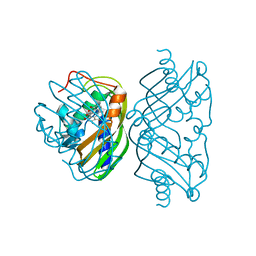 | | Structure of polyketide cyclase SnoaL | | Descriptor: | METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, nogalonic acid methyl ester cyclase | | Authors: | Sultana, A, Kallio, P, Jansson, A, Wang, J.S, Neimi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation.
Embo J., 23, 2004
|
|
2KZN
 
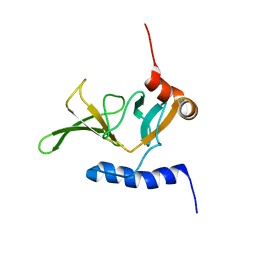 | | Solution NMR Structure of Peptide methionine sulfoxide reductase msrB from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR10 | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Ertekin, A, Maglaqui, M, Janjua, H, Cooper, B, Ciccosanti, C, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J, Lee, H, Aramini, J.M, Rossi, P, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1JGI
 
 | |
1NU5
 
 | |
3TXD
 
 | | HEWL co-crystallization with carboplatin in aqueous media with glycerol as the cryoprotectant | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Schreurs, A.M.M, Helliwell, J.R, Kroon-Batenburg, L.M.J. | | Deposit date: | 2011-09-23 | | Release date: | 2013-01-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Experience with exchange and archiving of raw data: comparison of data from two diffractometers and four software packages on a series of lysozyme crystals.
J.Appl.Crystallogr., 46, 2013
|
|
