6XH1
 
 | |
6I5H
 
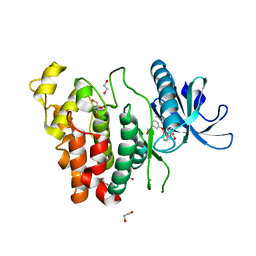 | | Crystal structure of CLK1 in complex with furanopyrimidin VN412 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-(3-phenoxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, ... | | Authors: | Schroeder, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
8XBG
 
 | |
5ZFZ
 
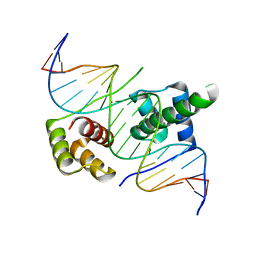 | | Crystal structure of human DUX4 homeodomains bound to A12T DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*TP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*AP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5Z9J
 
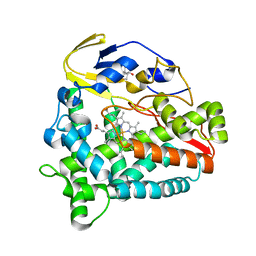 | | Identification of the functions of unusual cytochrome p450-like monooxygenases involved in microbial secondary metablism | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like enzyme, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lu, M, Lin, L, Zhang, C, Chen, Y. | | Deposit date: | 2018-02-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Riboflavin Is Directly Involved in the N-Dealkylation Catalyzed by Bacterial Cytochrome P450 Monooxygenases.
Chembiochem, 2020
|
|
6NV9
 
 | | BACE1 in complex with a macrocyclic inhibitor | | Descriptor: | (3S)-3-hydroxy-N-(2-methylpropyl)-N~2~-{[(4S)-17-[(methylsulfonyl)(propyl)amino]-2-oxo-3-azatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaen-4-yl]methyl}-L-norleucinamide, Beta-secretase 1, SULFATE ION | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-02-04 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Development of an Efficient Enzyme Production and Structure-Based Discovery Platform for BACE1 Inhibitors.
Biochemistry, 58, 2019
|
|
6NW3
 
 | | BACE1 in complex with a macrocyclic inhibitor | | Descriptor: | Beta-secretase 1, N-(2-methylpropyl)-N~2~-{[(4S)-17-[(methylsulfonyl)(propyl)amino]-2-oxo-3-azatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaen-4-yl]methyl}-L-norleucinamide, SULFATE ION | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-02-05 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Development of an Efficient Enzyme Production and Structure-Based Discovery Platform for BACE1 Inhibitors.
Biochemistry, 58, 2019
|
|
6I1C
 
 | | Crystal structure of Chlamydomonas reinhardtii thioredoxin f2 | | Descriptor: | thioredoxin f2 | | Authors: | Lemaire, S.D, Tedesco, D, Crozet, P, Michelet, L, Fermani, S, Zaffagnini, M, Henri, J. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Chloroplastic Thioredoxin f2 fromChlamydomonas reinhardtiiReveals Distinct Surface Properties.
Antioxidants (Basel), 7, 2018
|
|
5YV9
 
 | | Structure of CaMKK2 in complex with CKI-009 | | Descriptor: | 5-chloro-2-methoxy-4[(1Z)-3-(4-methoxyphenyl)-3-oxoprop-1-en-1-yl]aminobenzoic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
6I2H
 
 | |
6XLW
 
 | | Crystal structure of U2AF65 bound to AdML splice site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(P*UP*UP*(UD)P*UP*U)-D(P*(BRU))-R(P*CP*C)-3'), GLYCEROL, ... | | Authors: | Maji, D, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2020-06-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Representative cancer-associated U2AF2 mutations alter RNA interactions and splicing.
J.Biol.Chem., 295, 2020
|
|
5ZIQ
 
 | | Crystal structure of hexacoordinated heme protein from anhydrobiotic tardigrade at pH 4 | | Descriptor: | 1,2-ETHANEDIOL, Globin protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kim, J, Fukuda, Y, Inoue, T. | | Deposit date: | 2018-03-16 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Kumaglobin: a hexacoordinated heme protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus.
FEBS J., 286, 2019
|
|
6N3E
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to U2AF ligand motif 4 | | Descriptor: | FORMIC ACID, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
5ZLH
 
 | | Crystal structure of Mn-ProtoporphyrinIX-reconstituted P450BM3 | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, MANGANESE PROTOPORPHYRIN IX | | Authors: | Omura, K, Aiba, Y, Onoda, H, Sugimoto, H, Shoji, O, Watanabe, Y. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Chem. Commun. (Camb.), 54, 2018
|
|
8PI2
 
 | |
8F7U
 
 | | Macrocyclic Plasmin Inhibitor | | Descriptor: | (5S,8R,18S,21R)-N-{[4-(aminomethyl)phenyl]methyl}-21-[(benzenesulfonyl)amino]-3,11,20-trioxo-2,5,8,12,19-pentaazatetracyclo[21.2.2.2~5,8~.2~13,16~]hentriaconta-1(25),13,15,23,26,28-hexaene-18-carboxamide, Activation peptide, GLYCEROL, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-11-20 | | Release date: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Synthesis and structural characterization of new macrocyclicplasmin inhibitors
Chemmedchem, 2023
|
|
6XU5
 
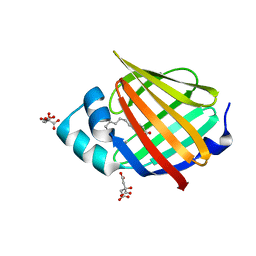 | | Human myelin protein P2 mutant N2D | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
8FXQ
 
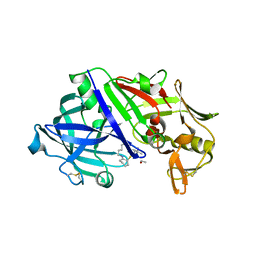 | | The Crystal Sturucture of Rhizopuspepsin with a bound modified peptide inhibitor generated by de novo drug design. | | Descriptor: | ALA-CYS-VAL-LYS, CYCLOHEXANE, Rhizopuspepsin, ... | | Authors: | Satyshur, K.A, Rich, D.H, Ripka, A.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Aspartic protease inhibitors designed from computer-generated templates bind as predicted.
Org Lett, 3, 2001
|
|
8PFV
 
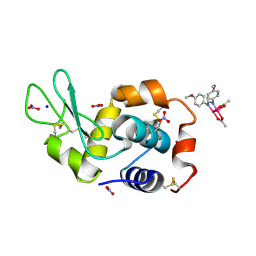 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DAniF)(O2CCH3)3] in condition A | | Descriptor: | 9,11-bis(4-methoxyphenyl)-3,7-dimethyl-2,4,6,8-tetraoxa-9,11-diaza-1$l^{4},5$l^{4}-diruthenatricyclo[3.3.3.0^{1,5}]undecane, CHLORIDE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
8PFU
 
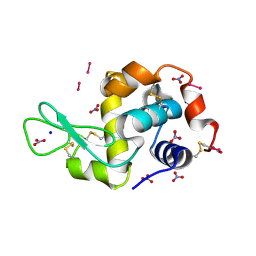 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K3[Ru2(CO3)4] in condition A | | Descriptor: | 6,8-bis(4-fluorophenyl)-1,5-bis(oxidanyl)-2,4-dioxa-6,8-diaza-1$l^{4},5$l^{4}-diruthenabicyclo[3.3.0]octan-3-one, CARBONATE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
8PFY
 
 | |
8PFT
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K2[Ru2(D-p-FPhF)(CO3)3] in condition A | | Descriptor: | 6,8-bis(4-fluorophenyl)-1,5-bis(oxidanyl)-2,4-dioxa-6,8-diaza-1$l^{4},5$l^{4}-diruthenabicyclo[3.3.0]octan-3-one, CHLORIDE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
5YZF
 
 | | X-ray crystal structure of met K42C sperm whale myoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, sperm whale myoglobin | | Authors: | Yuan, H. | | Deposit date: | 2017-12-14 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Formation of Cys-heme cross-link in K42C myoglobin under reductive conditions with molecular oxygen
J. Inorg. Biochem., 182, 2018
|
|
8F7V
 
 | | Macrocyclic plasmin inhibitor | | Descriptor: | (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(benzenesulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, GLYCEROL, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-11-20 | | Release date: | 2023-02-08 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthesis and structural characterization of new macrocyclicplasmin inhibitors
Chemmedchem, 2023
|
|
8PFW
 
 | |
