5VD6
 
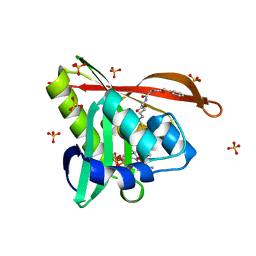 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 6 | | Descriptor: | (3R,5S,9R,23S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14-dioxo-23-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatetracosan-24-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
5IF3
 
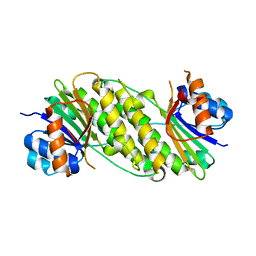 | |
7LSM
 
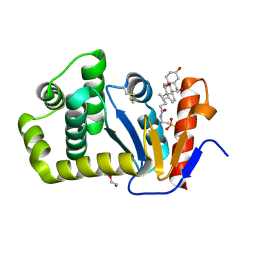 | | Crystal structure of E.coli DsbA in complex with bile salt taurocholate | | Descriptor: | DI(HYDROXYETHYL)ETHER, TAUROCHOLIC ACID, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Selective Binding of Small Molecules to Vibrio cholerae DsbA Offers a Starting Point for the Design of Novel Antibacterials.
Chemmedchem, 17, 2022
|
|
6VRO
 
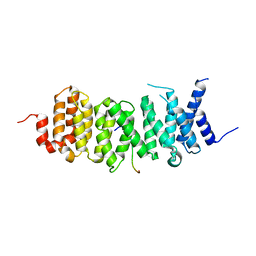 | | The structure of the PP2A B56 subunit AIM1 complex | | Descriptor: | Beta/gamma crystallin domain-containing protein 1, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2020-02-08 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A dynamic charge-charge interaction modulates PP2A:B56 substrate recruitment.
Elife, 9, 2020
|
|
5VDI
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Mutant N38Q Bound to AM-3607, Glycine, and Ivermectin | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Human GlyR alpha 3 Bound to Ivermectin.
Structure, 25, 2017
|
|
5IFC
 
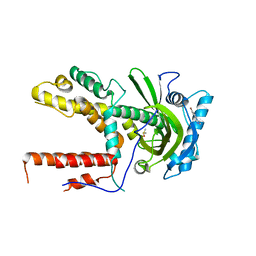 | |
5IFM
 
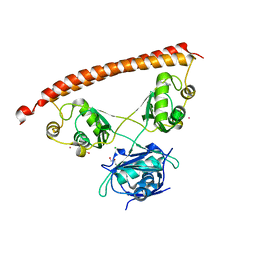 | | Human NONO (p54nrb) Homodimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-POU domain-containing octamer-binding protein, ... | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of human NONO (p54(nrb)): overcoming pathological problems with purification, data collection and noncrystallographic symmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8VUS
 
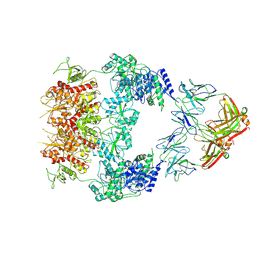 | | Human GluN1-2A with IgG 007-168 | | Descriptor: | 007-168 Heavy, 007-168 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5IOC
 
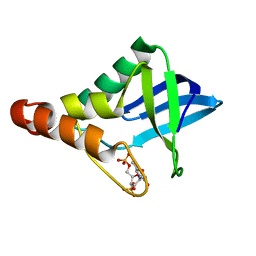 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66H/V99D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Theodoru, A, Garcia-Moreno E, B. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66H/V99D at cryogenic temperature
To be Published
|
|
8W14
 
 | |
6VTM
 
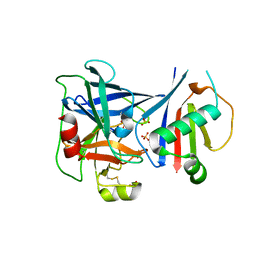 | |
8VUT
 
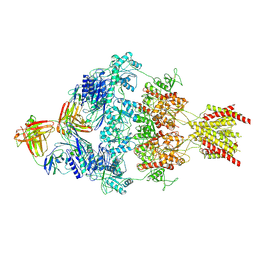 | | Human GluN1-2A with IgG 008-218 | | Descriptor: | 008-218 Heavy, 008-218 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5IOL
 
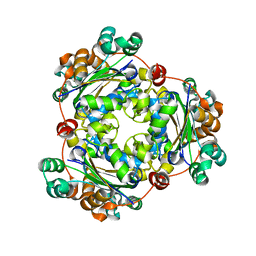 | | Crystal structure of Nucleoside Diphosphate Kinase from Schistosoma mansoni | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Torini, J.R.S, Romanello, L, Bird, L.E, Nettleship, J.E, Owens, R.J, Aller, P, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-03-08 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Characterization of a Schistosoma mansoni NDPK expressed in sexual and digestive organs.
Mol.Biochem.Parasitol., 2019
|
|
6VIO
 
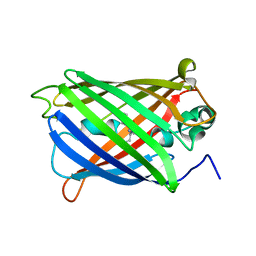 | |
5IPE
 
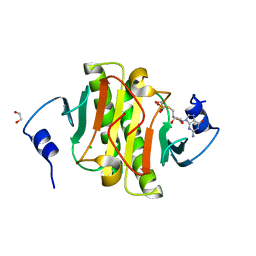 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) nucleoside thiophosphoramidate catalytic product complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-phosphono-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
8VTA
 
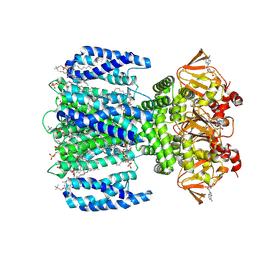 | | SthK R120A in the presence of PIP2 and cAMP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Transcriptional regulator, ... | | Authors: | Schmidpeter, P.A.M, Thon, O, Nimigean, C.M. | | Deposit date: | 2024-01-26 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | PIP2 inhibits pore opening of the cyclic nucleotide-gated channel SthK.
Nat Commun, 15, 2024
|
|
8VPI
 
 | |
7M5B
 
 | |
5VDR
 
 | |
5IGU
 
 | | Macrolide 2'-phosphotransferase type II | | Descriptor: | Macrolide 2'-phosphotransferase II | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2016-02-28 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Kinase-Mediated Macrolide Antibiotic Resistance.
Structure, 25, 2017
|
|
5IH0
 
 | |
8VK8
 
 | | Structure of UbV.d2.1 in complex with Ube2d2 | | Descriptor: | Ubiquitin variant D2.1, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Middleton, A.J. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-25 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural and biophysical characterisation of ubiquitin variants that inhibit the ubiquitin conjugating enzyme Ube2d2.
Febs J., 291, 2024
|
|
8VVH
 
 | | rat GluN1a-2B Fab 003-102 local refinement | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-31 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5IHF
 
 | |
6VU9
 
 | |
