6V8B
 
 | |
6W45
 
 | | Crystal structure of HAO1 in complex with biaryl acid inhibitor - compound 3 | | Descriptor: | 2-chloranyl-4-[2-[[(6-chloranyl-1~{H}-indol-2-yl)carbonyl-methyl-amino]methyl]-5-fluoranyl-phenyl]benzoic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W44
 
 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 4 | | Descriptor: | 5-[methyl-[(2-propoxypyridin-3-yl)methyl]amino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W4C
 
 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 5 | | Descriptor: | 5-[[3-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-2-oxidanyl-phenyl]methylamino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
1VJI
 
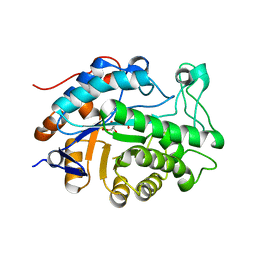 | | Gene Product of At1g76680 from Arabidopsis thaliana | | Descriptor: | 12-oxophytodienoate reductase (OPR1), FLAVIN MONONUCLEOTIDE | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | X-ray structure of Arabidopsis At1g77680, 12-oxophytodienoate reductase isoform 1.
Proteins, 61, 2005
|
|
6A6V
 
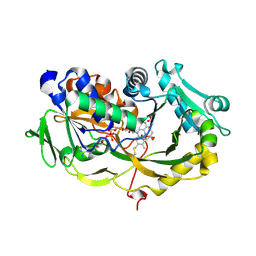 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with 7 additional mutations, in complex with FSA | | Descriptor: | 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6R
 
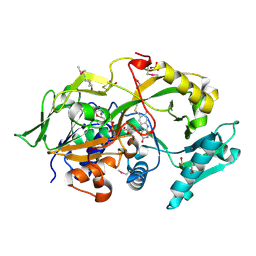 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6ARR
 
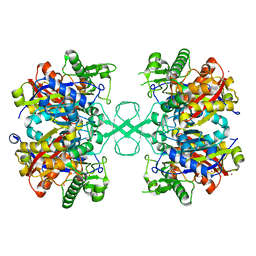 | | Aspergillus fumigatus Cytosolic Thiolase: Apo enzyme in complex with cesium ions | | Descriptor: | Acetyl-CoA acetyltransferase, CESIUM ION, CHLORIDE ION, ... | | Authors: | Marshall, A.C, Bond, C.S, Bruning, J.B. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Structure of Aspergillus fumigatus Cytosolic Thiolase: Trapped Tetrahedral Reaction Intermediates and Activation by Monovalent Cations
Acs Catalysis, 8(3), 2018
|
|
7A56
 
 | | Schmallenberg Virus Envelope Glycoprotein Gc Fusion Domains in Postfusion Conformation | | Descriptor: | CHLORIDE ION, Envelopment polyprotein, PHOSPHATE ION, ... | | Authors: | Hellert, J, Guardado-Calvo, P, Rey, F.A. | | Deposit date: | 2020-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure, function, and evolution of the Orthobunyavirus membrane fusion glycoprotein.
Cell Rep, 42, 2023
|
|
6AQP
 
 | | Aspergillus fumigatus Cytosolic Thiolase: Acetylated enzyme in complex with CoA and potassium ions | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA acetyltransferase, CHLORIDE ION, ... | | Authors: | Marshall, A.C, Bond, C.S, Bruning, J.B. | | Deposit date: | 2017-08-21 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Aspergillus fumigatus Cytosolic Thiolase: Trapped Tetrahedral Reaction Intermediates and Activation by Monovalent Cations
Acs Catalysis, 8(3), 2018
|
|
7AW9
 
 | | CCAAT-binding complex and HapX bound to Aspergillus fumigatus cccA DNA | | Descriptor: | BZIP domain-containing protein, CBFD_NFYB_HMF domain-containing protein, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2020-11-06 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX.
Structure, 30, 2022
|
|
1SRX
 
 | |
6ARF
 
 | | Aspergillus fumigatus Cytosolic Thiolase: Apo enzyme in complex with potassium ions | | Descriptor: | Acetyl-CoA acetyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marshall, A.C, Bond, C.S, Bruning, J.B. | | Deposit date: | 2017-08-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structure of Aspergillus fumigatus Cytosolic Thiolase: Trapped Tetrahedral Reaction Intermediates and Activation by Monovalent Cations
Acs Catalysis, 8(3), 2018
|
|
6ARG
 
 | | Aspergillus fumigatus Cytosolic Thiolase: Apo enzyme in complex with rubidium ions | | Descriptor: | Acetyl-CoA acetyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marshall, A.C, Bond, C.S, Bruning, J.B. | | Deposit date: | 2017-08-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Structure of Aspergillus fumigatus Cytosolic Thiolase: Trapped Tetrahedral Reaction Intermediates and Activation by Monovalent Cations
Acs Catalysis, 8(3), 2018
|
|
6ART
 
 | | Aspergillus fumigatus Cytosolic Thiolase: Apo enzyme in complex with cesium ions | | Descriptor: | Acetyl-CoA acetyltransferase, CESIUM ION, CHLORIDE ION, ... | | Authors: | Marshall, A.C, Bond, C.S, Bruning, J.B. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Aspergillus fumigatus Cytosolic Thiolase: Trapped Tetrahedral Reaction Intermediates and Activation by Monovalent Cations
Acs Catalysis, 8(3), 2018
|
|
6H3U
 
 | | Schmallenberg Virus Glycoprotein Gc Head Domain in Complex with scFv 4B6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hellert, J, Aebischer, A, Wernike, K, Haouz, A, Brocchi, E, Reiche, S, Guardado-Calvo, P, Beer, M, Rey, F.A. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.168 Å) | | Cite: | Orthobunyavirus spike architecture and recognition by neutralizing antibodies.
Nat Commun, 10, 2019
|
|
6H3T
 
 | | Schmallenberg Virus Glycoprotein Gc Head Domain in Complex with scFv 1C11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hellert, J, Aebischer, A, Wernike, K, Haouz, A, Brocchi, E, Reiche, S, Guardado-Calvo, P, Beer, M, Rey, F.A. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Orthobunyavirus spike architecture and recognition by neutralizing antibodies.
Nat Commun, 10, 2019
|
|
6H3S
 
 | | Schmallenberg Virus Glycoprotein Gc Head/Stalk Domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Envelopment polyprotein, ... | | Authors: | Hellert, J, Aebischer, A, Wernike, K, Haouz, A, Brocchi, E, Reiche, S, Guardado-Calvo, P, Beer, M, Rey, F.A. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | Orthobunyavirus spike architecture and recognition by neutralizing antibodies.
Nat Commun, 10, 2019
|
|
6IC4
 
 | |
6RS8
 
 | | X-ray crystal structure of LsAA9B (transition metals soak) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
7U0U
 
 | | Crystal Structure of a Aspergillus fumigatus Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fox III, D, Abendroth, J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
6RS7
 
 | | X-ray crystal structure of LsAA9B (deglycosylated form) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AA9, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
3ZHF
 
 | | gamma 2 adaptin EAR domain crystal structure with preS1 site1 peptide NPDWDFN | | Descriptor: | 1,2-ETHANEDIOL, AP-1 COMPLEX SUBUNIT GAMMA-LIKE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Juergens, M.C, Voros, J, Rautureau, G, Shepherd, D, Pye, V.E, Muldoon, J, Johnson, C.M, Ashcroft, A, Freund, S.M.V, Ferguson, N. | | Deposit date: | 2012-12-21 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Hepatitis B Virus Pres1 Domain Hijacks Host Trafficking Proteins by Motif Mimicry.
Nat.Chem.Biol., 9, 2013
|
|
6RS9
 
 | | X-ray crystal structure of LsAA9B (xylotetraose soak) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AA9, BICINE, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
7LS9
 
 | |
