2JLT
 
 | | Crystal structure of an RNA kissing complex | | Descriptor: | R06, TAR | | Authors: | DiPrimo, C, Fribourg, S. | | Deposit date: | 2008-09-15 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exploring Tar-RNA Aptamer Loop-Loop Interaction by X-Ray Crystallography, Uv Spectroscopy and Surface Plasmon Resonance.
Nucleic Acids Res., 36, 2008
|
|
315D
 
 | |
1FIX
 
 | |
8EEY
 
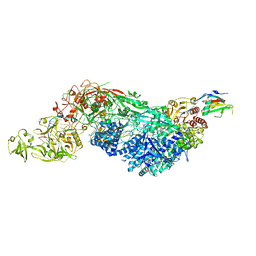 | | Cas7-11 in complex with DR-mismatched target RNA, Csx29 and Csx30 | | Descriptor: | Cas7-11, Csx29, Csx30, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
1AXP
 
 | | DNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 6 STRUCTURES | | Descriptor: | DNA (D(GAAGAGAAGC)(DOT)D(GCTTCTCTTC)) | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-17 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
7WFX
 
 | | EVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-27 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
165D
 
 | | THE STRUCTURE OF A MISPAIRED RNA DOUBLE HELIX AT 1.6 ANGSTROMS RESOLUTION AND IMPLICATIONS FOR THE PREDICTION OF RNA SECONDARY STRUCTURE | | Descriptor: | DNA/RNA (5'-R(*GP*CP*UP*UP*CP*GP*GP*CP*)-D(*(BRU))-3'), RHODIUM HEXAMINE ION | | Authors: | Cruse, W, Saludjian, P, Biala, E, Strazewski, P, Prange, T, Kennard, O. | | Deposit date: | 1994-03-21 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a mispaired RNA double helix at 1.6-A resolution and implications for the prediction of RNA secondary structure.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
3WNC
 
 | | Crystal structure of EF-Pyl in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
1E6M
 
 | | TWO-COMPONENT SIGNAL TRANSDUCTION SYSTEM D57A MUTANT OF CHEY | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
4H5P
 
 | |
1E6K
 
 | | Two-component signal transduction system D12A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
1E6L
 
 | | Two-component signal transduction system D13A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
4YCW
 
 | | Crystal structure of cladosporin in complex with plasmodium like human lysyl-tRNA synthetase mutant | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Fang, P, Wang, J, Guo, M. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Specific Inhibition of tRNA Synthetase by ATP Competitive Inhibitor
Chem.Biol., 22, 2015
|
|
7WG4
 
 | | DVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-28 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WG2
 
 | | EVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-28 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WG1
 
 | | DVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-27 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8OQI
 
 | | Cryo-EM structure of the wild-type alpha-synuclein fibril. | | Descriptor: | Alpha-synuclein | | Authors: | Pesch, V, Reithofer, S, Ma, L, Flores-Fernandez, J.M, Oezduezenciler, P, Busch, Y, Lien, Y, Rudtke, O, Frieg, B, Schroeder, G.F, Wille, H, Tamgueney, G. | | Deposit date: | 2023-04-12 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Vaccination with structurally adapted fungal protein fibrils induces immunity to Parkinson's disease.
Brain, 147, 2024
|
|
2GV4
 
 | | Solution structure of the poliovirus 3'-UTR Y-stem | | Descriptor: | Short RNA strand 5'-GGACCUCUCGAAAGAGUGGUCC-3' | | Authors: | Heus, H.A, Zoll, J, Tessari, M, van Kuppeveld, F.J.M, Melchers, W.J.G. | | Deposit date: | 2006-05-02 | | Release date: | 2007-03-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Breaking pseudo-twofold symmetry in the poliovirus 3'-UTR Y-stem by restoring Watson-Crick base pairs.
Rna, 13, 2007
|
|
168D
 
 | |
8EDJ
 
 | | Crystal structure of rA3G-ssRNA-GA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*GP*AP*UP*UP*U)-3'), SULFATE ION, ... | | Authors: | Pacheco, J, Yang, H.J, Li, S.-X, Chen, X.S. | | Deposit date: | 2022-09-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
3LJE
 
 | | The X-ray structure of zebrafish RNase5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase5 | | Authors: | Russo Krauss, I, Merlino, A, Coscia, F, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-11-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
3LJD
 
 | | The X-ray structure of zebrafish RNase1 from a new crystal form at pH 4.5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase1 | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
6ZK7
 
 | | Crystal Structure of human PYROXD1/FAD complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulfide oxidoreductase domain-containing protein 1 | | Authors: | Meinhart, A, Asanovic, I, Martinez, J, Clausen, T. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The oxidoreductase PYROXD1 uses NAD(P) + as an antioxidant to sustain tRNA ligase activity in pre-tRNA splicing and unfolded protein response.
Mol.Cell, 81, 2021
|
|
2LBW
 
 | | Solution structure of the S. cerevisiae H/ACA RNP protein Nhp2p-S82W mutant | | Descriptor: | H/ACA ribonucleoprotein complex subunit 2 | | Authors: | Koo, B, Park, C, Fernandez, C.F, Chim, N, Ding, Y, Chanfreau, G, Feigon, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of H/ACA RNP Protein Nhp2p Reveals Cis/Trans Isomerization of a Conserved Proline at the RNA and Nop10 Binding Interface.
J.Mol.Biol., 411, 2011
|
|
2LBX
 
 | | Solution structure of the S. cerevisiae H/ACA RNP protein Nhp2p | | Descriptor: | H/ACA ribonucleoprotein complex subunit 2 | | Authors: | Koo, B, Park, C, Fernandez, C.F, Chim, N, Ding, Y, Chanfreau, G, Feigon, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of H/ACA RNP Protein Nhp2p Reveals Cis/Trans Isomerization of a Conserved Proline at the RNA and Nop10 Binding Interface.
J.Mol.Biol., 411, 2011
|
|
