2FRB
 
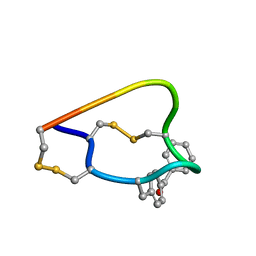 | | NMR structure of the alpha-conotoxin GI (ASN4)-benzoylphenylalanine derivative | | Descriptor: | Alpha-conotoxin GIA | | Authors: | Pashkov, V.S, Maslennikov, I, Kasheverov, I.V, Zhmak, M.N, Utkin, Y.N, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 2006-01-19 | | Release date: | 2006-05-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Alpha-Conotoxin GI benzoylphenylalanine derivatives. (1)H-NMR structures and photoaffinity labeling of the Torpedo californica nicotinic acetylcholine receptor.
Febs J., 273, 2006
|
|
3JQ4
 
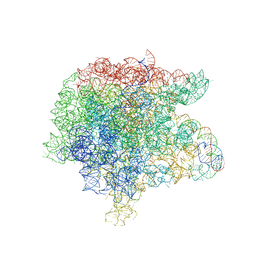 | | The structure of the complex of the large ribosomal subunit from D. Radiodurans with the antibiotic lankacidin | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, N-[(1S,2R,3E,5E,7S,9E,11E,13S,15R,19R)-7,13-dihydroxy-1,4,10,19-tetramethyl-17,18-dioxo-16-oxabicyclo[13.2.2]nonadeca-3,5,9,11-tetraen-2-yl]-2-oxopropanamide | | Authors: | Auerbach-Nevo, T, Mermershtain, I, Davidovich, C, Bashan, A, Rozenberg, H, Yonath, A. | | Deposit date: | 2009-09-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The structure of ribosome-lankacidin complex reveals ribosomal sites for synergistic antibiotics
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8HR4
 
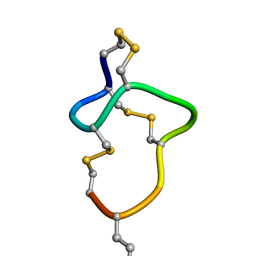 | | [D-Cys5,D-Lys16]-STp(5-17) | | Descriptor: | DCY-CYS-GLU-LEU-CYS-CYS-ASN-PRO-ALA-CYS-ALA-DLY-CYS | | Authors: | Shimamoto, S, Hidaka, Y, Yoshino, S, Goto, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The molecular basis of heat-stable enterotoxin for vaccine development and cancer cell detection
To Be Published
|
|
6NES
 
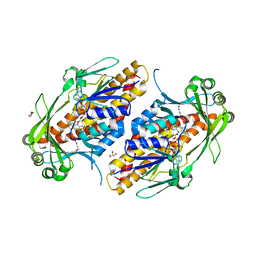 | | FAD-dependent monooxygenase TropB from T. stipitatus | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6ZE1
 
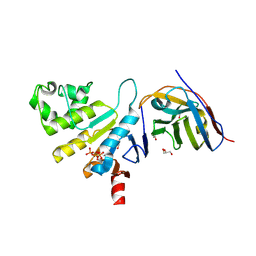 | | human NBD1 of CFTR in complex with nanobody G11a | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Garcia-Pino, A, Govaerts, C, Scholl, D, Sigoillot, M. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A topological switch in CFTR modulates channel activity and sensitivity to unfolding.
Nat.Chem.Biol., 17, 2021
|
|
6XAA
 
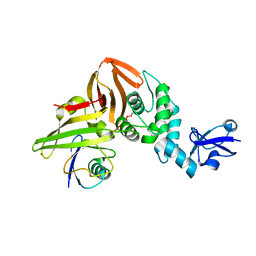 | | SARS CoV-2 PLpro in complex with ubiquitin propargylamide | | Descriptor: | Non-structural protein 3, Ubiquitin-propargylamide, ZINC ION | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6P4E
 
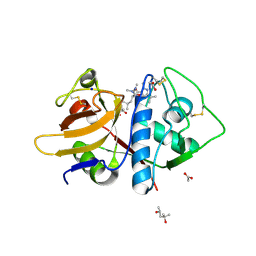 | | Leishmania mexicana CPB in complex with an aza-nitrile inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-tert-butyl-N-[(2S)-1-(2-cyano-1,2-dimethylhydrazinyl)-4-methyl-1-oxopentan-2-yl]-1-methyl-1H-pyrazole-5-carboxamide (non-preferred name), ... | | Authors: | Ribeiro, J.F.R, Li, C, De Vita, D, Emsley, J, Montanari, C.A. | | Deposit date: | 2019-05-27 | | Release date: | 2020-06-03 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution X-Ray Crystal Structures of LmCPB2.8 Co-Crystalized with Dipeptidyl Aza-nitrile Inhibitor and Structure Activity Relationships
To Be Published
|
|
5JVF
 
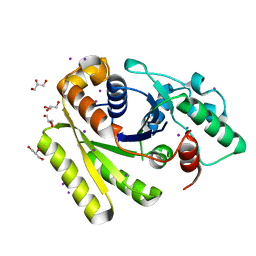 | | Crystal Structure of Apo-FleN | | Descriptor: | GLYCEROL, IODIDE ION, Site-determining protein | | Authors: | Jain, D, Chanchal, Banerjee, P. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | ATP-Induced Structural Remodeling in the Antiactivator FleN Enables Formation of the Functional Dimeric Form
Structure, 25, 2017
|
|
5J4N
 
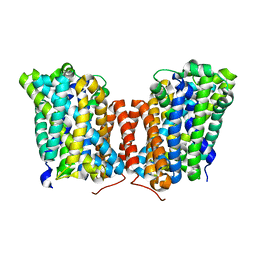 | |
1GMM
 
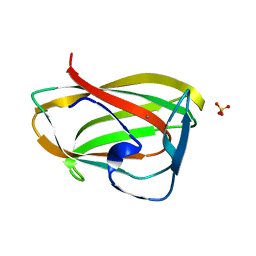 | | Carbohydrate binding module CBM6 from xylanase U Clostridium thermocellum | | Descriptor: | CALCIUM ION, CBM6, SODIUM ION, ... | | Authors: | Czjzek, M, Mosbah, A, Bolam, D, Allouch, J, Zamboni, V, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2001-09-19 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Location of the Ligand-Binding Site of Carbohydrate-Binding Modules that Have Evolved from a Common Sequence is not Conserved.
J.Biol.Chem., 276, 2001
|
|
6ZHL
 
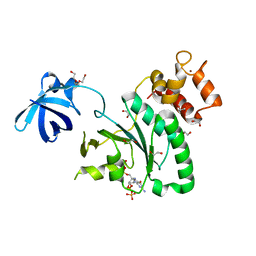 | | Crystal Structure of Staphylococcus aureus RsgA bound to ppGpp. | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, Small ribosomal subunit biogenesis GTPase RsgA, ... | | Authors: | Bennison, D.J, Rafferty, J.B, Corrigan, R.M. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Stringent Response Inhibits 70S Ribosome Formation in Staphylococcus aureus by Impeding GTPase-Ribosome Interactions.
Mbio, 12, 2021
|
|
9FUD
 
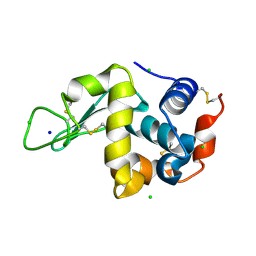 | | Serial microseconds crystallography at ID29 using fixed-target (Si Chip): Lysozyme - without ligand GlcNAc (apo) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Orlans, J, Rose, S.L, Basu, S, de Sanctis, D. | | Deposit date: | 2024-06-26 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Advancing macromolecular structure determination with microsecond X-ray pulses at a 4th generation synchrotron.
Commun Chem, 8, 2025
|
|
5JIP
 
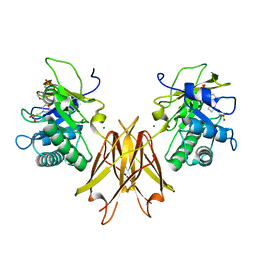 | | Crystal structure of the Clostridium perfringens spore cortex lytic enzyme SleM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cortical-lytic enzyme, MAGNESIUM ION | | Authors: | Chirgadze, D.Y, Christie, G, Ustok, F.I, Al-Riyami, B, Stott, K. | | Deposit date: | 2016-04-22 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Clostridium perfringens SleM, a muramidase involved in cortical hydrolysis during spore germination.
Proteins, 84, 2016
|
|
8AAC
 
 | |
7M0R
 
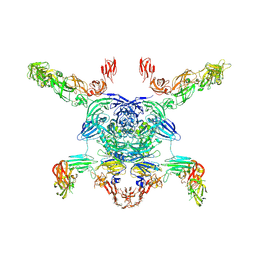 | | Cryo-EM structure of the Sema3A/PlexinA4/Neuropilin 1 complex | | Descriptor: | CALCIUM ION, Neuropilin-1, Plexin-A4, ... | | Authors: | Lu, D, Shang, G, He, X, Bai, X, Zhang, X. | | Deposit date: | 2021-03-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the Sema3A/PlexinA4/Neuropilin tripartite complex.
Nat Commun, 12, 2021
|
|
5J5S
 
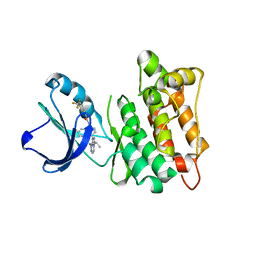 | | Src kinase in complex with a sulfonamide inhibitor | | Descriptor: | N-{4-[8-amino-3-(propan-2-yl)imidazo[1,5-a]pyrazin-1-yl]naphthalen-1-yl}-N'-[3-(trifluoromethyl)phenyl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Lebedev, I, Feldman, H.C, Georghiou, G, Maly, D.J, Seeliger, M. | | Deposit date: | 2016-04-03 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Structural and Functional Analysis of the Allosteric Modulation of IRE1a with ATP-Competitive Kinase Inhibitors
To Be Published
|
|
9N1T
 
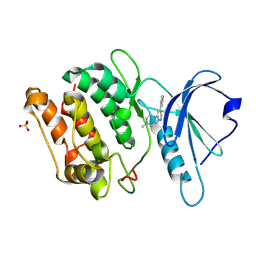 | |
2BBI
 
 | |
5LWV
 
 | | Human OGT in complex with UDP and fused substrate peptide (HCF1) | | Descriptor: | GLYCEROL, Host cell factor 1,UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, PHOSPHATE ION, ... | | Authors: | Raimi, O, Rafie, K, Kapuria, V, Herr, W, van Aalten, D. | | Deposit date: | 2016-09-19 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats.
Open Biol, 7, 2017
|
|
7M3P
 
 | | Xrcc4-Spc110p(164-207) fusion | | Descriptor: | Xrcc4-Spc110p(164-207) | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.0000186 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
2W2R
 
 | | Structure of the vesicular stomatitis virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
7ZYD
 
 | | Structure of Compound 6 Bound to CK2alpha | | Descriptor: | 5-bromanyl-6-chloranyl-1~{H}-indole, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-24 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
7TUD
 
 | | Crystal structure of HLA-B*44:05 (T73C) with 6mer EEFGRC and dipeptide GL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, EEFGRC peptide, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7MMY
 
 | |
1GMX
 
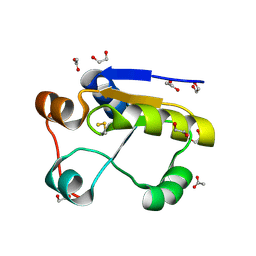 | | Escherichia coli GlpE sulfurtransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
