6X2F
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - L2 state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X43
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - II state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X4W
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - III state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X50
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - V state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewelyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X4Y
 
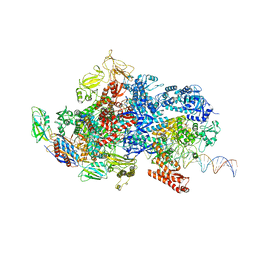 | | Mfd-bound E.coli RNA polymerase elongation complex - IV state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6XEO
 
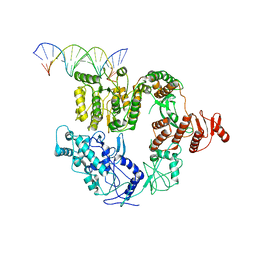 | | Structure of Mfd bound to dsDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*AP*TP*AP*CP*TP*TP*AP*CP*AP*GP*CP*CP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*GP*GP*CP*TP*GP*TP*AP*AP*GP*TP*AP*TP*CP*CP*T)-3'), Transcription-repair-coupling factor | | Authors: | Brugger, C, Deaconescu, A. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd.
Nat Commun, 11, 2020
|
|
6XKI
 
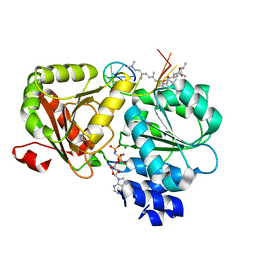 | | Crystal structure of eIF4A-I in complex with RNA bound to des-MePateA, a pateamine A analog | | Descriptor: | (3S,6Z,8E,11S,15R)-15-amino-3-[(1E,3E,5E)-7-(dimethylamino)-2,5-dimethylhepta-1,3,5-trien-1-yl]-9,11-dimethyl-4,12-dioxa-20-thia-21-azabicyclo[16.2.1]henicosa-1(21),6,8,18-tetraene-5,13-dione, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Liang, J, Naineni, S.K, Pelletier, J, Nagar, B. | | Deposit date: | 2020-06-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Functional mimicry revealed by the crystal structure of an eIF4A:RNA complex bound to the interfacial inhibitor, desmethyl pateamine A.
Cell Chem Biol, 28, 2021
|
|
6ZM2
 
 | |
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6ZNS
 
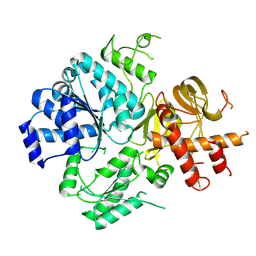 | | Crystal Structure of DUF1998 helicase MrfA | | Descriptor: | Uncharacterized ATP-dependent helicase YprA, ZINC ION | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
6ZNP
 
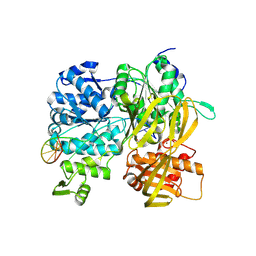 | | Crystal Structure of DUF1998 helicase MrfA bound to DNA | | Descriptor: | CITRIC ACID, Uncharacterized ATP-dependent helicase YprA, ZINC ION, ... | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
6ZNQ
 
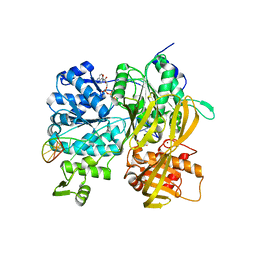 | | Crystal Structure of DUF1998 helicase MrfA bound to DNA and AMPPNP | | Descriptor: | CITRIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized ATP-dependent helicase YprA, ... | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
6ZQE
 
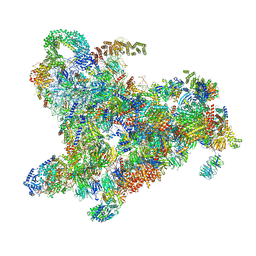 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-A (Poly-Ala) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQG
 
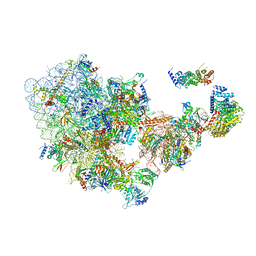 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-C | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQD
 
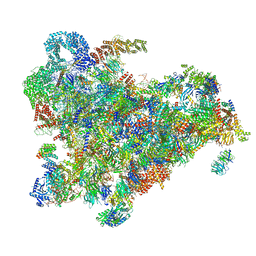 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Post-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQF
 
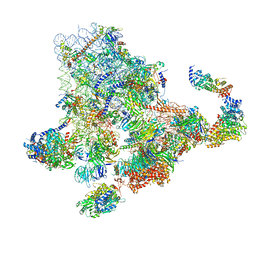 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-B (Poly-Ala) | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZWW
 
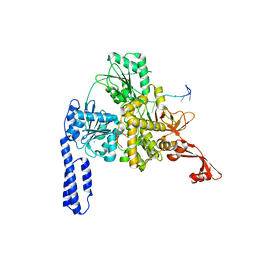 | | Crystal structure of E. coli RNA helicase HrpA in complex with RNA | | Descriptor: | ATP-dependent RNA helicase HrpA, CALCIUM ION, ssRNA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZWX
 
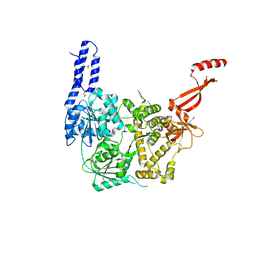 | | Crystal structure of E. coli RNA helicase HrpA | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JL3
 
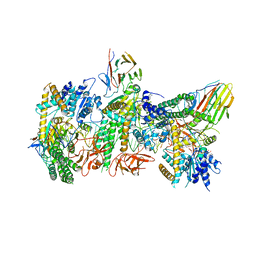 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL1
 
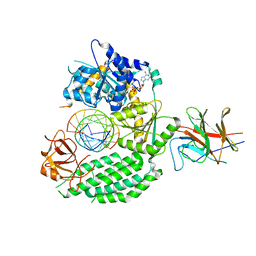 | | Cryo-EM structure of RIG-I:dsRNA in complex with RIPLET PrySpry domain (monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
6ZYM
 
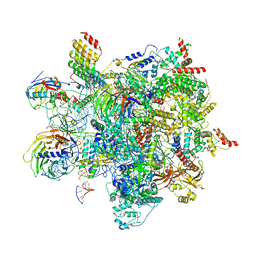 | | Human C Complex Spliceosome - High-resolution CORE | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Corepressor interacting with RBPJ 1, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
7A5P
 
 | | Human C Complex Spliceosome - Medium-resolution PERIPHERY | | Descriptor: | Cell division cycle 5-like protein, Crooked neck-like protein 1, Eukaryotic initiation factor 4A-III, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
7A8R
 
 | | Structure of RecQL from Bos taurus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase, MAGNESIUM ION, ... | | Authors: | Rety, S, Chen, W.F, Xi, X.G. | | Deposit date: | 2020-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Endogenous Bos taurus RECQL is predominantly monomeric and more active than oligomers.
Cell Rep, 36, 2021
|
|
7ABG
 
 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ABI
 
 | | Human pre-Bact-2 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Beta-catenin-like protein 1, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
