1J51
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM MUTANT (F87W/Y96F/V247L/C334A) WITH 1,3,5-TRICHLOROBENZENE | | Descriptor: | 1,3,5-TRICHLORO-BENZENE, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Chen, X, Christopher, A, Jones, J, Guo, Q, Xu, F, Cao, R, Wong, L.L, Rao, Z. | | Deposit date: | 2002-01-05 | | Release date: | 2002-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the F87W/Y96F/V247L mutant of cytochrome P-450cam with 1,3,5-trichlorobenzene bound and further protein engineering for the oxidation of pentachlorobenzene and hexachlorobenzene
J.BIOL.CHEM., 277, 2002
|
|
6W8I
 
 | | Ternary complex structure - BTK cIAP compound 15 | | Descriptor: | 14-{[(3S)-2-(N-methyl-L-alanyl-3-methyl-L-valyl)-3-{[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]carbamoyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}-3,6,9,12-tetraoxatetradecan-1-yl (3R)-3-{5-amino-4-carbamoyl-3-[4-(2,4-difluorophenoxy)phenyl]-1H-pyrazol-1-yl}piperidine-1-carboxylate, Baculoviral IAP repeat-containing protein 2, Tyrosine-protein kinase BTK, ... | | Authors: | Calabrese, M.F, Schiemer, J.S. | | Deposit date: | 2020-03-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Characterization of BTK:PROTAC:cIAP Ternary Complexes: From Snapshots to Ensembles
Nat.Chem.Biol., 2020
|
|
1J86
 
 | | HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), MONOCLINIC CRYSTAL FORM 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garman, S.C, Sechi, S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2001-05-20 | | Release date: | 2001-08-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The analysis of the human high affinity IgE receptor Fc epsilon Ri alpha from multiple crystal forms.
J.Mol.Biol., 311, 2001
|
|
7OER
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-(2,2-diphenylethyl)-1,5-dimethyl-N-(2-(methylamino)-2-oxoethyl)-6-oxo-1,6-dihydropyridine-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-(2,2-diphenylethyl)-1,5-dimethyl-N-[2-(methylamino)-2-oxidanylidene-ethyl]-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of a Highly Selective BET BD2 Inhibitor from a DNA-Encoded Library Technology Screening Hit.
J.Med.Chem., 64, 2021
|
|
7OES
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 1,5-dimethyl-N-(2-(methylamino)-2-oxoethyl)-6-oxo-N-(2-phenylpropyl)-1,6-dihydropyridine-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 1,5-dimethyl-N-[2-(methylamino)-2-oxidanylidene-ethyl]-6-oxidanylidene-N-[(2S)-2-phenylpropyl]pyridine-3-carboxamide, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of a Highly Selective BET BD2 Inhibitor from a DNA-Encoded Library Technology Screening Hit.
J.Med.Chem., 64, 2021
|
|
2BSS
 
 | | Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B2705 | | Descriptor: | BETA-2-MICROGLOBULIN, HIV PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN B-27 ALPHA CHAIN | | Authors: | Stewart-Jones, G.B.E, Di Gleria, K, Kollnberger, S, Mcmichael, A.J, Jones, E.Y, Bowness, P. | | Deposit date: | 2005-05-23 | | Release date: | 2005-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures and Kir3Dl1 Recognition of Three Immunodominant Viral Peptides Complexed to Hla-B2705
Eur.J.Immunol., 35, 2005
|
|
1IDS
 
 | | X-RAY STRUCTURE ANALYSIS OF THE IRON-DEPENDENT SUPEROXIDE DISMUTASE FROM MYCOBACTERIUM TUBERCULOSIS AT 2.0 ANGSTROMS RESOLUTIONS REVEALS NOVEL DIMER-DIMER INTERACTIONS | | Descriptor: | FE (III) ION, IRON SUPEROXIDE DISMUTASE | | Authors: | Cooper, J.B, Mcintyre, K, Wood, S.P, Zhang, Y, Young, D. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of the iron-dependent superoxide dismutase from Mycobacterium tuberculosis at 2.0 Angstroms resolution reveals novel dimer-dimer interactions.
J.Mol.Biol., 246, 1995
|
|
2RQU
 
 | | Solution structure of the complex between the DDEF1 SH3 domain and the APC SAMP1 motif | | Descriptor: | 19-mer from Adenomatous polyposis coli protein, DDEF1_SH3 | | Authors: | Kaieda, S, Matsui, C, Mimori-Kiyosue, Y, Ikegami, T. | | Deposit date: | 2009-12-14 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the recognition of the SAMP motif of adenomatous polyposis coli by the Src-homology 3 domain.
Biochemistry, 49, 2010
|
|
1J02
 
 | | Crystal Structure of Rat Heme Oxygenase-1-Heme Bound to NO | | Descriptor: | HEME OXYGENASE 1, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2002-10-28 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
2W2M
 
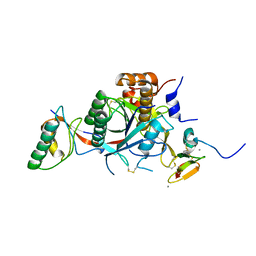 | | WT PCSK9-DELTAC BOUND TO WT EGF-A OF LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
7NRR
 
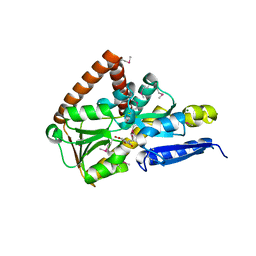 | | The structure of the SBP TarP_Csal in complex with caffeate | | Descriptor: | CAFFEIC ACID, MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-04 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NR2
 
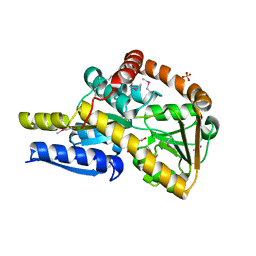 | | The structure of the SBP TarP_Sse in complex with coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NQG
 
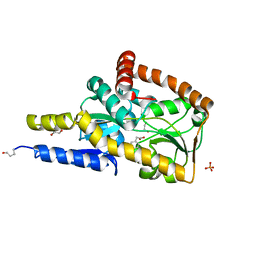 | | The structure of the SBP TarP_Rhp in complex with 4-hydroxyphenylacetate | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENYLACETATE, PHOSPHATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NRA
 
 | | The structure of the SBP TarP_Sse in complex with cinnamate | | Descriptor: | HYDROCINNAMIC ACID, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NSW
 
 | | The structure of the SBP TarP_Csal in complex with coumarate | | Descriptor: | 1,2-ETHANEDIOL, 4'-HYDROXYCINNAMIC ACID, MAGNESIUM ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NTD
 
 | | The structure of the SBP TarP_Csal in complex with ferulate | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NTE
 
 | | The structure of an open conformation of the SBP TarP_Csal | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
2W2N
 
 | | WT PCSK9-deltaC bound to EGF-A H306Y mutant of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2ULL
 
 | |
2REE
 
 | |
1IPP
 
 | | HOMING ENDONUCLEASE/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED ENDONUCLEASE I-PPOI, ... | | Authors: | Flick, K.E, Jurica, M.S, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 1998-03-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI.
Nature, 394, 1998
|
|
2RVA
 
 | |
2RV9
 
 | |
1J7I
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa Apoenzyme | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1J7M
 
 | | The Third Fibronectin Type II Module from Human Matrix Metalloproteinase 2 | | Descriptor: | MATRIX METALLOPROTEINASE 2 | | Authors: | Briknarova, K, Gehrmann, M, Banyai, L, Tordai, H, Patthy, L, Llinas, M. | | Deposit date: | 2001-05-17 | | Release date: | 2001-05-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Gelatin-binding region of human matrix metalloproteinase-2: solution structure, dynamics, and function of the COL-23 two-domain construct.
J.Biol.Chem., 276, 2001
|
|
