1YFG
 
 | | YEAST INITIATOR TRNA | | Descriptor: | YEAST INITIATOR TRNA | | Authors: | Basavappa, R, Sigler, P.B. | | Deposit date: | 1997-05-08 | | Release date: | 1997-09-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination.
EMBO J., 10, 1991
|
|
4Z7K
 
 | |
7PTK
 
 | |
4MSR
 
 | | RNA 10mer duplex with six 2'-5'-linkages | | Descriptor: | RNA 10mer duplex with six 2'-5'-linkages, STRONTIUM ION | | Authors: | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3QRR
 
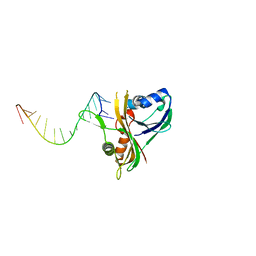 | | Structure of Thermus Thermophilus Cse3 bound to an RNA representing a product complex | | Descriptor: | Putative uncharacterized protein TTHB192, RNA (5'-R(*GP*UP*CP*CP*CP*CP*AP*CP*GP*CP*GP*UP*GP*UP*GP*GP*GP*(23G))-3') | | Authors: | Schellenberg, M.J, Gesner, E.G, Garside, E.L, MacMillan, A.M. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Recognition and maturation of effector RNAs in a CRISPR interference pathway.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4MSB
 
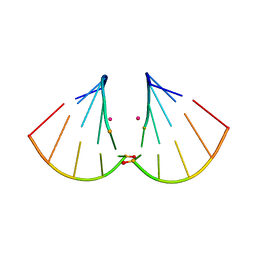 | | RNA 10mer duplex with two 2'-5'-linkages | | Descriptor: | RNA 10mer duplex with two 2'-5'-linkages, STRONTIUM ION | | Authors: | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4JRI
 
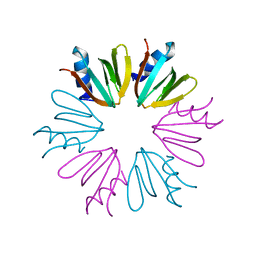 | |
1YRJ
 
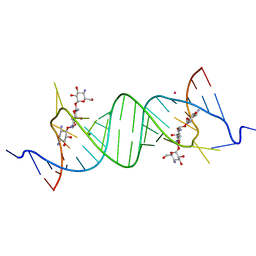 | | Crystal Structure of Apramycin bound to a Ribosomal RNA A site oligonucleotide | | Descriptor: | APRAMYCIN, Bacterial 16 S Ribosomal RNA A Site Oligonucleotide, MAGNESIUM ION, ... | | Authors: | Han, Q, Zhao, Q, Fish, S, Simonsen, K.B, Vourloumis, D, Froelich, J.M, Wall, D, Hermann, T. | | Deposit date: | 2005-02-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition by glycoside pseudo base pairs and triples in an apramycin-RNA complex.
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
4JLI
 
 | |
4JUV
 
 | |
5VH7
 
 | | Structure and dynamics of RNA repeat expansions that cause Huntington's Disease and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*CP*AP*GP*CP*AP*G)-3') | | Authors: | Chen, J.L, VanEtten, D.M, Fountain, M.A, Yildirim, I, Disney, M.D. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of RNA Repeat Expansions That Cause Huntington's Disease and Myotonic Dystrophy Type 1.
Biochemistry, 56, 2017
|
|
5VH8
 
 | | Structure and dynamics of RNA repeat expansions that cause Huntington's Disease and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*CP*UP*GP*CP*UP*GP*CP*UP*GP*GP*UP*C)-3') | | Authors: | Chen, J.L, VanEtten, D.M, Fountain, M.A, Yildirim, I, Disney, M.D. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of RNA Repeat Expansions That Cause Huntington's Disease and Myotonic Dystrophy Type 1.
Biochemistry, 56, 2017
|
|
3O6E
 
 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1FT8
 
 | |
3QRQ
 
 | | Structure of Thermus Thermophilus Cse3 bound to an RNA representing a pre-cleavage complex | | Descriptor: | Putative uncharacterized protein TTHB192, RNA (5'-R(*GP*UP*CP*CP*CP*CP*AP*CP*GP*CP*GP*UP*GP*UP*GP*GP*GP*GP*A)-3') | | Authors: | Schellenberg, M.J, Gesner, E.G, Garside, E.L, MacMillan, A.M. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Recognition and maturation of effector RNAs in a CRISPR interference pathway.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3O7V
 
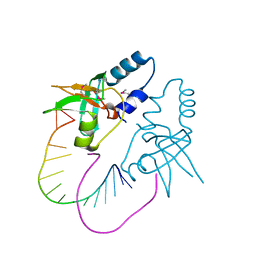 | | Crystal Structure of human Hiwi1 (V361M) PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-08-01 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4JRK
 
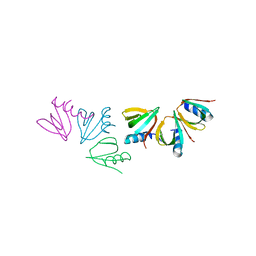 | |
2R8S
 
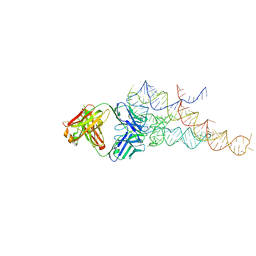 | | High resolution structure of a specific synthetic FAB bound to P4-P6 RNA ribozyme domain | | Descriptor: | Fab heavy chain, Fab light chain, MAGNESIUM ION, ... | | Authors: | Ye, J.D, Tereshko, V, Sidhu, S.S, Koide, S, Kossiakoff, A.A, Piccirilli, J.A. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthetic antibodies for specific recognition and crystallization of structured RNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2LBW
 
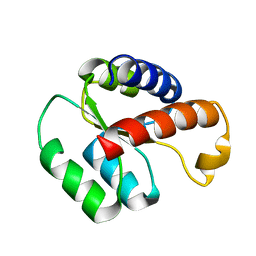 | | Solution structure of the S. cerevisiae H/ACA RNP protein Nhp2p-S82W mutant | | Descriptor: | H/ACA ribonucleoprotein complex subunit 2 | | Authors: | Koo, B, Park, C, Fernandez, C.F, Chim, N, Ding, Y, Chanfreau, G, Feigon, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of H/ACA RNP Protein Nhp2p Reveals Cis/Trans Isomerization of a Conserved Proline at the RNA and Nop10 Binding Interface.
J.Mol.Biol., 411, 2011
|
|
2LBX
 
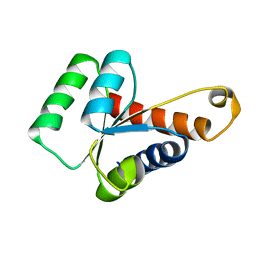 | | Solution structure of the S. cerevisiae H/ACA RNP protein Nhp2p | | Descriptor: | H/ACA ribonucleoprotein complex subunit 2 | | Authors: | Koo, B, Park, C, Fernandez, C.F, Chim, N, Ding, Y, Chanfreau, G, Feigon, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of H/ACA RNP Protein Nhp2p Reveals Cis/Trans Isomerization of a Conserved Proline at the RNA and Nop10 Binding Interface.
J.Mol.Biol., 411, 2011
|
|
248D
 
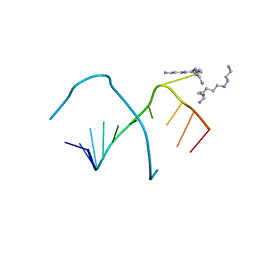 | | CRYSTAL STRUCTURES OF AN A-FORM DUPLEX WITH SINGLE-ADENOSINE BULGES AND A CONFORMATIONAL BASIS FOR SITE SPECIFIC RNA SELF-CLEAVAGE | | Descriptor: | DNA/RNA (5'-R(*GP*CP*GP*)-D(*AP*TP*AP*TP*AP*)-R(*CP*GP*C)-3'), ORTHORHOMBIC, SPERMINE | | Authors: | Portmann, S, Grimm, S, Workman, C, Usman, N, Egli, M. | | Deposit date: | 1996-02-02 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of an A-form duplex with single-adenosine bulges and a conformational basis for site-specific RNA self-cleavage.
Chem.Biol., 3, 1996
|
|
468D
 
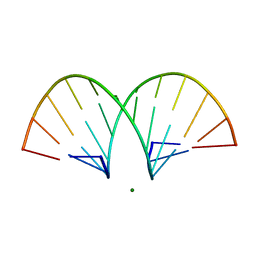 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
470D
 
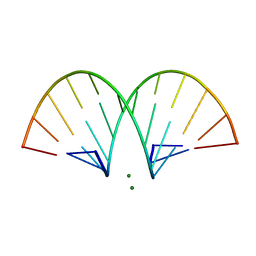 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
471D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
7ZHH
 
 | | Complex structure of drosophila Unr CSD789 and a poly(A) RNA sequence | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, Upstream of N-ras, ... | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res., 51, 2023
|
|
