7JPX
 
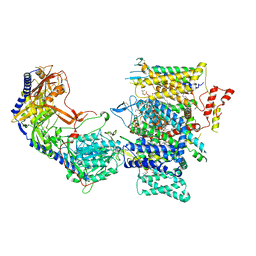 | | Rabbit Cav1.1 in the presence of 100 micromolar amlodipine in nanodiscs at 2.9 Angstrom resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-08-10 | | Release date: | 2020-11-18 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca v 1.1 by Dihydropyridine Compounds*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5H9Y
 
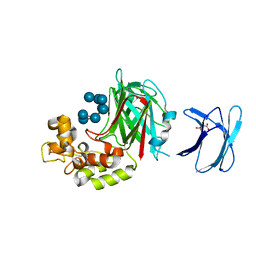 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii complexed with laminarihexaose. | | Descriptor: | L(+)-TARTARIC ACID, beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
6BW8
 
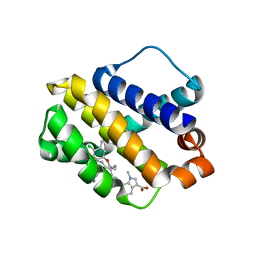 | | Mcl-1 complexed with small molecules | | Descriptor: | 7-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-3-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Optimization of Potent and Selective Tricyclic Indole Diazepinone Myeloid Cell Leukemia-1 Inhibitors Using Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6F9R
 
 | | Crystal structure of human Angiotensin-1 converting enzyme N-domain in complex with Sampatrilat-Asp. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of sampatrilat and sampatrilat-Asp in complex with human ACE - a molecular basis for domain selectivity.
FEBS J., 285, 2018
|
|
6F9T
 
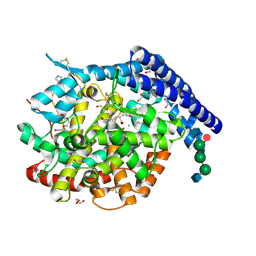 | | Crystal structure of human testis Angiotensin-1 converting enzyme in complex with Sampatrilat. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of sampatrilat and sampatrilat-Asp in complex with human ACE - a molecular basis for domain selectivity.
FEBS J., 285, 2018
|
|
6BHJ
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | Descriptor: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
2YIP
 
 | |
4CLM
 
 | | Structure of Salmonella typhi type I dehydroquinase irreversibly inhibited with a 1,3,4-trihydroxyciclohexane-1-carboxylic acid derivative | | Descriptor: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Tizon, L, Maneiro, M, Lence, E, Poza, S, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Irreversible covalent modification of type I dehydroquinase with a stable Schiff base.
Org. Biomol. Chem., 13, 2015
|
|
2OTY
 
 | | 1,2-dichlorobenzene in complex with T4 Lysozyme L99A | | Descriptor: | 1,2-DICHLOROBENZENE, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Graves, A.P, Shoichet, B.K. | | Deposit date: | 2007-02-09 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Predicting absolute ligand binding free energies to a simple model site.
J.Mol.Biol., 371, 2007
|
|
1OMZ
 
 | | crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2),
a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
2OU0
 
 | |
3ANN
 
 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with quinolin-2-ylmethylphosphonic acid | | Descriptor: | (quinolin-2-ylmethyl)phosphonic acid, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Endo, K, Kato, M, Deng, L, Song, Y, Yajima, S. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase/Lipophilic Phosphonate Complexes
ACS Med Chem Lett, 2, 2011
|
|
4CEX
 
 | | 1.59 A resolution Fluoride inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, FLUORIDE ION, NICKEL (II) ION, ... | | Authors: | Benini, S, Cianci, M, Ciurli, S. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Fluoride Inhibition of Sporosarcina Pasteurii Urease: Structure and Thermodynamics.
J.Biol.Inorg.Chem., 19, 2014
|
|
3AEC
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-(1-methylethyl)-benzamid | | Descriptor: | 2-iodo-N-(1-methylethyl)benzamide, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-(1-methylethyl)-benzamid
To be Published
|
|
4CEU
 
 | | 1.58 A resolution native Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Benini, S, Cianci, M, Ciurli, S. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Fluoride Inhibition of Sporosarcina Pasteurii Urease: Structure and Thermodynamics.
J.Biol.Inorg.Chem., 19, 2014
|
|
2CZD
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution
To be Published
|
|
1XSE
 
 | | Crystal Structure of Guinea Pig 11beta-Hydroxysteroid Dehydrogenase Type 1 | | Descriptor: | 11beta-hydroxysteroid dehydrogenase type 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogg, D, Elleby, B, Norstrom, C, Stefansson, K, Abrahmsen, L, Oppermann, U, Svensson, S. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of guinea pig 11beta-hydroxysteroid dehydrogenase type 1 provides a model for enzyme-lipid bilayer interactions
J.Biol.Chem., 280, 2005
|
|
4HI9
 
 | | 1.2 structure of integrin-linked kinase ankyrin repeat domain in complex with PINCH1 LIM1 domain collected at wavelength 0.91974 | | Descriptor: | IODIDE ION, Integrin-linked protein kinase, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Stiegler, A.L, Jakoncic, J, Stojanoff, V, Chiswell, B.P, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-10-11 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.203 Å) | | Cite: | 1.2 structure of integrin-linked kinase ankyrin repeat domain in complex with PINCH1 LIM1 domain collected at wavelength 0.91974
To be Published
|
|
1QAT
 
 | |
1QFO
 
 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH 3'SIALYLLACTOSE | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, PROTEIN (SIALOADHESIN) | | Authors: | May, A.P, Robinson, R.C, Vinson, M, Crocker, P.R, Jones, E.Y. | | Deposit date: | 1999-04-12 | | Release date: | 1999-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the N-terminal domain of sialoadhesin in complex with 3' sialyllactose at 1.85 A resolution.
Mol.Cell, 1, 1998
|
|
3AH3
 
 | | Crystal structure of LR5-1, 3-isopropylmalate dehydrogenase created by directed evolution | | Descriptor: | 1,2-ETHANEDIOL, Homoisocitrate dehydrogenase, SULFATE ION | | Authors: | Tomita, T, Suzuki, Y, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2010-04-13 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enhancement of the latent 3-isopropylmalate dehydrogenase activity of promiscuous homoisocitrate dehydrogenase by directed evolution
Biochem.J., 431, 2010
|
|
7CX8
 
 | | Structure of the CYP102A1 Haem Domain with N-(5-Cyclohexyl)valeroyl-L-Phenylalanine in complex with (R)-1-Tetralylamine | | Descriptor: | (1R)-1,2,3,4-tetrahydronaphthalen-1-amine, (1~{S})-1,2,3,4-tetrahydronaphthalen-1-amine, (2~{S})-2-(5-cyclohexylpentanoylamino)-3-phenyl-propanoic acid, ... | | Authors: | Stanfield, J.K, Sugimoto, H, Shoji, O. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the CYP102A1 Haem Domain with N-(5-Cyclohexyl)valeroyl-L-Phenylalanine in complex with (R)-1-Tetralylamine at 1.70 Angstrom Resolution
To Be Published
|
|
6I8T
 
 | |
1AEJ
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (1-VINYLIMIDAZOLE) | | Descriptor: | 1-VINYLIMIDAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
6A7B
 
 | | AKR1C3 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C3, DIMETHYLFORMAMIDE, ... | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
