7MD6
 
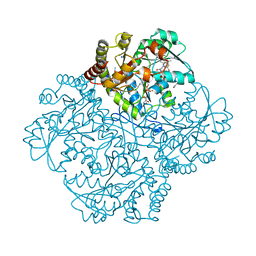 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103N mutant co-crystallized with NL1 | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, CITRATE ANION, N-[(6-bromo-1H-indol-1-yl)acetyl]glycine, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
2GO2
 
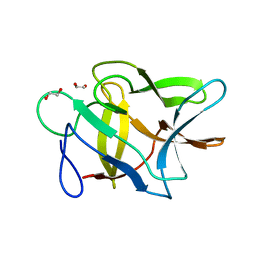 | |
5N86
 
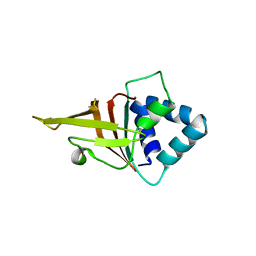 | | Crystal structure of FAS1 domain of hyaluronic acid receptor stabilin-2 | | Descriptor: | Stabilin-2 | | Authors: | Twarda-Clapa, A, Labuzek, B, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Crystal structure of the FAS1 domain of the hyaluronic acid receptor stabilin-2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3IZL
 
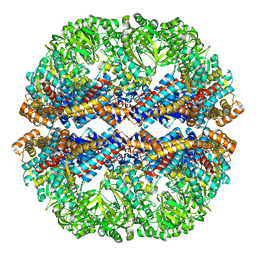 | | Mm-cpn rls deltalid with ATP and AlFx | | Descriptor: | Mm-cpn rls deltalid | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3B7D
 
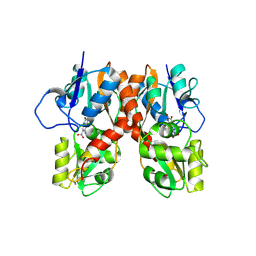 | |
7MDB
 
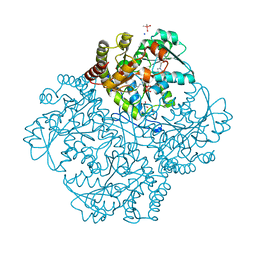 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103A mutant co-crystallized with NL2 | | Descriptor: | 5-[(6-bromo-1H-indol-1-yl)methyl]-2-methylfuran-3-carboxylic acid, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, CACODYLATE ION, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
2NUY
 
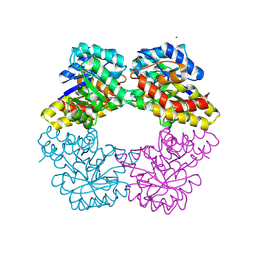 | |
3J1B
 
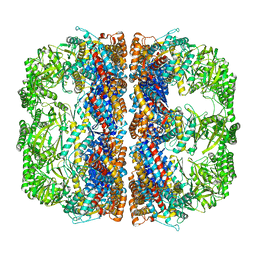 | | Cryo-EM structure of 8-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
7M8S
 
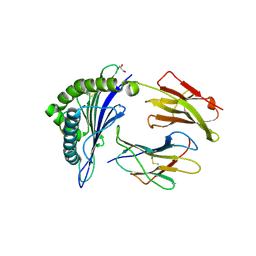 | | Crystal Structure of HLA-A*02:01 in complex with KLNDLCFTNV, an 10-mer epitope from SARS-CoV-2 Spike (S386-395) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | SARS-CoV-2 Spike-Derived Peptides Presented by HLA Molecules
Biophysica, 1, 2021
|
|
2NZI
 
 | |
7MCN
 
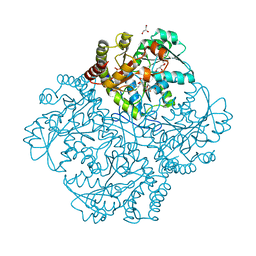 | | Crystal structure of Staphylococcus aureus Cystathionine gamma-lyase, Holoenzyme with High HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MD0
 
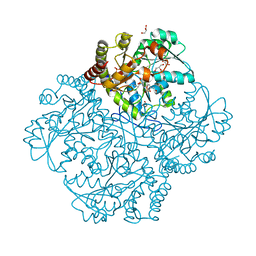 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme in the presence of NL1F3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7FD5
 
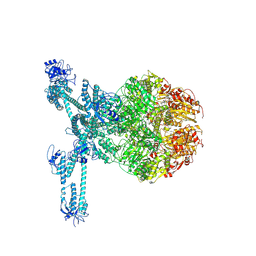 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
3J03
 
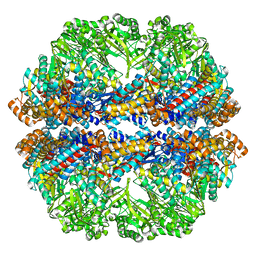 | | Lidless Mm-cpn in the closed state with ATP/AlFx | | Descriptor: | Lidless Mm-cpn | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
3J1C
 
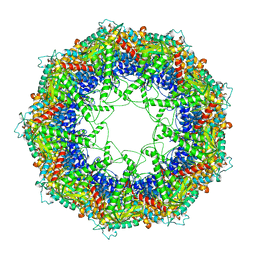 | | Cryo-EM structure of 9-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3BG1
 
 | | Architecture of a Coat for the Nuclear Pore Membrane | | Descriptor: | Nucleoporin NUP145, Protein SEC13 homolog | | Authors: | Hoelz, A. | | Deposit date: | 2007-11-23 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architecture of a coat for the nuclear pore membrane.
Cell(Cambridge,Mass.), 131, 2007
|
|
7MCT
 
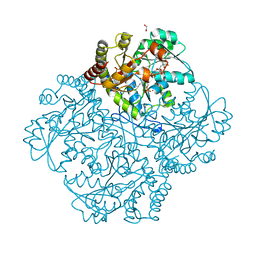 | | Crystal structure of Staphylococcus aureus Cystathionine gamma lyase, Holoenzyme with bound NL1 | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, N-[(6-bromo-1H-indol-1-yl)acetyl]glycine, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
2H0J
 
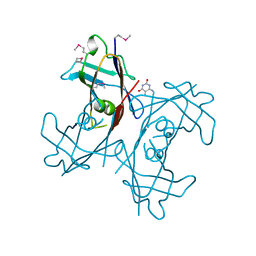 | | Crystal structure of PucM in the presence of 5,6-diaminouracil | | Descriptor: | 5,6-DIAMINOPYRIMIDINE-2,4(1H,3H)-DIONE, Transthyretin-like protein pucM | | Authors: | Rhee, S. | | Deposit date: | 2006-05-15 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional analysis of PucM, a hydrolase in the ureide pathway and a member of the transthyretin-related protein family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5N1N
 
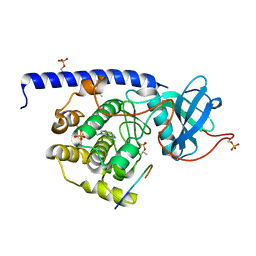 | |
3IZK
 
 | | Mm-cpn rls deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
7MTO
 
 | |
7EPW
 
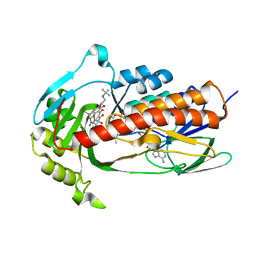 | | Crystal structure of monooxygenase Tet(X4) with tigecycline | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, TIGECYCLINE | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2021-04-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline.
Bmc Biol., 19, 2021
|
|
7MCL
 
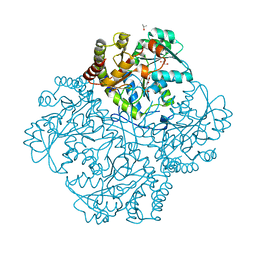 | | Crystal structure of Staphylococcus aureus Cystathionine gamma-lyase, PLP bound | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
2H0W
 
 | |
3BAM
 
 | |
