3KRB
 
 | |
3ANR
 
 | | human DYRK1A/harmine complex | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Nonaka, Y, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2010-09-06 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A
Nat Commun, 1, 2010
|
|
1ICS
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1PRH
 
 | |
3HVN
 
 | | Crystal structure of cytotoxin protein suilysin from Streptococcus suis | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, HEPTANE-1,2,3-TRIOL, Hemolysin | | Authors: | Xu, L, Huang, B, Du, H, Zhang, C.X, Xu, J, Li, X, Rao, Z. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Crystal structure of cytotoxin protein suilysin from Streptococcus suis.
Protein Cell, 1, 2010
|
|
6BLQ
 
 | | Crystal Structure of IAg7 in complex with insulin mimotope p8E9E | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BLX
 
 | | Crystal structure of IAg7 in complex with insulin mimotope p8G9E | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3SXV
 
 | | Crystal structure of the complex of goat lactoperoxidase with amitrole at 2.1 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with amitrole at 2.1 A resolution
TO BE PUBLISHED
|
|
5FTG
 
 | | Human choline kinase a1 in complex with compound 1-[[4-[2-[4-[[4-(dimethylamino)pyridin-1- yl]methyl]phenoxy]ethoxy]phenyl]methyl]-N,N- dimethyl-pyridin-4-amine (compound 10a) | | Descriptor: | 1,2-ETHANEDIOL, 1-[[4-[2-[4-[[4-(dimethylamino)pyridin-1-yl]methyl]phenoxy]ethoxy]phenyl]methyl]-N,N-dimethyl-pyridin-4-amine, CHOLINE KINASE ALPHA | | Authors: | Schiaffino-Ortega, S, Baglioni, E, Mariotto, E, Bortolozzi, R, Serran-Aguilera, L, Rios-Marco, P, Carrasco-Jimenez, M.P, Gallo, M.A, Hurtado-Guerrero, R, Marco, C, Basso, G, Viola, G, Entrena, A, Lopez-Cara, L.C. | | Deposit date: | 2016-01-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design, Synthesis, Crystallization and Biological Evaluation of New Symmetrical Biscationic Compounds as Selective Inhibitors of Human Choline Kinase Alpha1 (Chokalpha1)
Sci.Rep., 6, 2016
|
|
4EJJ
 
 | | Human Cytochrome P450 2A6 in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2012-04-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone binding and access channel in human cytochrome P450 2A6 and 2A13 enzymes.
J.Biol.Chem., 287, 2012
|
|
7Y5O
 
 | | Crystal structure of human CAF-1 core complex in spacegroup P21 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5L
 
 | | Crystal structure of human CAF-1 core complex in spacegroup C2 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
3KLB
 
 | |
4CLD
 
 | |
4CM7
 
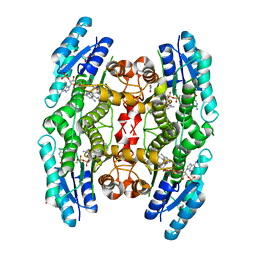 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 5-phenethyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
2OD6
 
 | |
1XAS
 
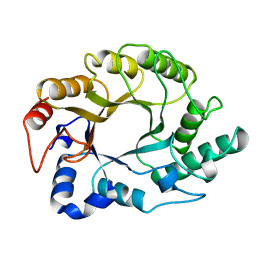 | | CRYSTAL STRUCTURE, AT 2.6 ANGSTROMS RESOLUTION, OF THE STREPTOMYCES LIVIDANS XYLANASE A, A MEMBER OF THE F FAMILY OF BETA-1,4-D-GLYCANSES | | Descriptor: | 1,4-BETA-D-XYLAN XYLANOHYDROLASE | | Authors: | Derewenda, U, Derewenda, Z.S. | | Deposit date: | 1994-05-31 | | Release date: | 1995-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure, at 2.6-A resolution, of the Streptomyces lividans xylanase A, a member of the F family of beta-1,4-D-glycanases.
J.Biol.Chem., 269, 1994
|
|
3LC7
 
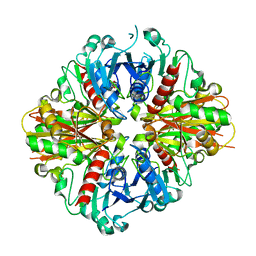 | | Crystal Structure of apo Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) from methicllin resistant Staphylococcus aureus (MRSA252) | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase 1 | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-01-10 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
1YBV
 
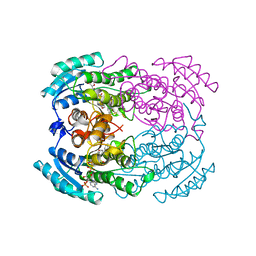 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5-METHYL-1,2,4-TRIAZOLO[3,4-B]BENZOTHIAZOLE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Andersson, A, Schneider, G, Lindqvist, Y. | | Deposit date: | 1996-09-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the ternary complex of 1,3,8-trihydroxynaphthalene reductase from Magnaporthe grisea with NADPH and an active-site inhibitor.
Structure, 4, 1996
|
|
3D4O
 
 | |
3O76
 
 | |
2A27
 
 | | Human DRP-1 kinase, W305S S308A D40 mutant, crystal form with 8 monomers in the asymmetric unit | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Death-associated protein kinase 2 | | Authors: | Kursula, P, Lehmann, F, Shani, G, Kimchi, A, Wilmanns, M. | | Deposit date: | 2005-06-22 | | Release date: | 2006-10-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DRP-1 kinase, W305S S308A D40 mutant, crystal form with 8 monomers in the asymmetric unit
To be Published
|
|
4EJG
 
 | | Human Cytochrome P450 2A13 in complex with Nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450 2A13, GLYCEROL, ... | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2012-04-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone binding and access channel in human cytochrome P450 2A6 and 2A13 enzymes.
J.Biol.Chem., 287, 2012
|
|
4EML
 
 | |
3LC2
 
 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from methicillin resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-01-09 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
