3TZ6
 
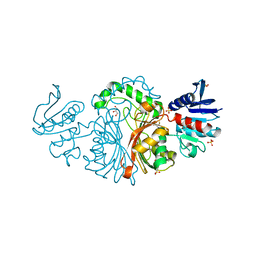 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With inhibitor SMCS (CYS) And Phosphate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CYSTEINE, GLYCEROL, ... | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1N82
 
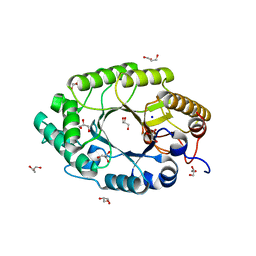 | | The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus | | Descriptor: | GLYCEROL, SODIUM ION, intra-cellular xylanase | | Authors: | Solomon, V, Teplitsky, A, Golan, G, Gilboa, R, Reiland, V, Shulami, S, Moryles, S, Zolotnitsky, G, Shoham, Y, Shoham, G. | | Deposit date: | 2002-11-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The high-resolution crystal structure of IXT6,
a thermophilic, intracellular xylanase from G. stearothermophilus
To be Published
|
|
3TXB
 
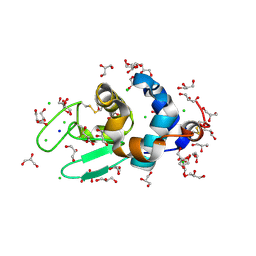 | | HEWL co-crystallization with cisplatin in aqueous media with glycerol as the cryoprotectant | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Schreurs, A.M.M, Helliwell, J.R, Kroon-Batenburg, L.M.J. | | Deposit date: | 2011-09-23 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Experience with exchange and archiving of raw data: comparison of data from two diffractometers and four software packages on a series of lysozyme crystals.
J.Appl.Crystallogr., 46, 2013
|
|
3TXJ
 
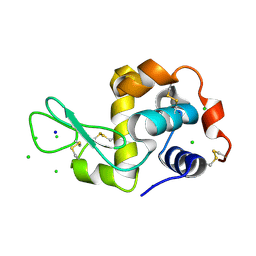 | | HEWL co-crystallization with NAG with silicone oil as the cryoprotectant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Schreurs, A.M.M, Helliwell, J.R, Kroon-Batenburg, L.M.J. | | Deposit date: | 2011-09-23 | | Release date: | 2013-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Experience with exchange and archiving of raw data: comparison of data from two diffractometers and four software packages on a series of lysozyme crystals.
J.Appl.Crystallogr., 46, 2013
|
|
3TZI
 
 | |
4E8C
 
 | | Crystal structure of streptococcal beta-galactosidase in complex with galactose | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35, ... | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
3U0V
 
 | | Crystal Structure Analysis of human LYPLAL1 | | Descriptor: | Lysophospholipase-like protein 1 | | Authors: | Burger, M, Zimmermann, T.J, Kondoh, Y, Stege, P, Watanabe, N, Osada, H, Waldmann, H, Vetter, I.R. | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases
J.Lipid Res., 53, 2012
|
|
3U37
 
 | |
4ETB
 
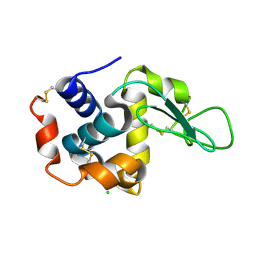 | | lysozyme, room temperature, 200 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4EKR
 
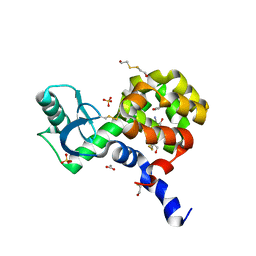 | | T4 Lysozyme L99A/M102H with 2-Cyanophenol Bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 2-hydroxybenzonitrile, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering a model protein cavity to catalyze the Kemp elimination.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3U6L
 
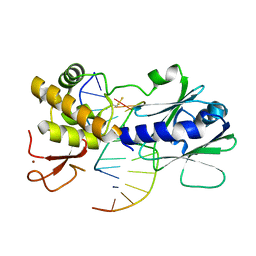 | | MutM set 2 CpGo | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*GP*TP*(CX2)P*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U57
 
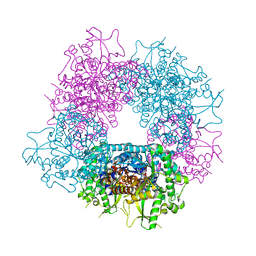 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | (2beta,7beta,16S,17R,19E,21beta)-21-(beta-D-glucopyranosyloxy)-2,7-dihydro-7,17-cyclosarpagan-17-yl acetate, CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
2YJN
 
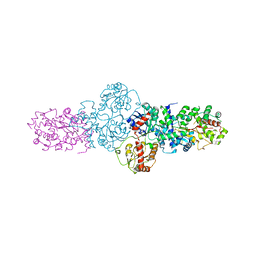 | | Structure of the glycosyltransferase EryCIII from the erythromycin biosynthetic pathway, in complex with its activating partner, EryCII | | Descriptor: | DTDP-4-KETO-6-DEOXY-HEXOSE 3,4-ISOMERASE, GLYCOSYLTRANSFERASE | | Authors: | Moncrieffe, M.C, Fernandez, M.J, Spiteller, D, Matsumura, H, Gay, N.J, Luisi, B.F, Leadlay, P.F. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Structure of the Glycosyltransferase Eryciii in Complex with its Activating P450 Homologue Erycii.
J.Mol.Biol., 415, 2012
|
|
3U5Y
 
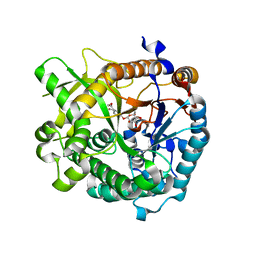 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, Secologanin | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
4EMJ
 
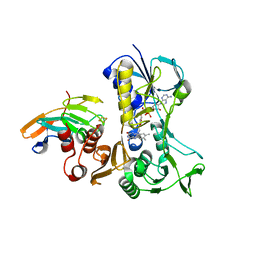 | | Complex between the reductase and ferredoxin components of toluene dioxygenase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, TodA, ... | | Authors: | Lin, T.Y, Werther, T, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2012-04-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Suppression of Electron Transfer to Dioxygen by Charge Transfer and Electron Transfer Complexes in the FAD-dependent Reductase Component of Toluene Dioxygenase.
J.Biol.Chem., 287, 2012
|
|
1N8Q
 
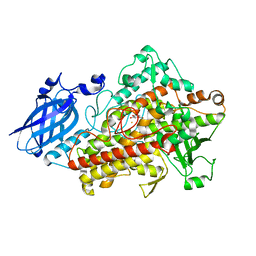 | | LIPOXYGENASE IN COMPLEX WITH PROTOCATECHUIC ACID | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (II) ION, lipoxygenase-3 | | Authors: | Borbulevych, O.Y, Jankun, J, Selman, S.H, Skrzypczak-Jankun, E. | | Deposit date: | 2002-11-21 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipoxygenase interactions with natural flavonoid, quercetin, reveal a complex with protocatechuic acid in its X-ray structure at 2.1 A resolution.
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
1LYZ
 
 | |
1LZA
 
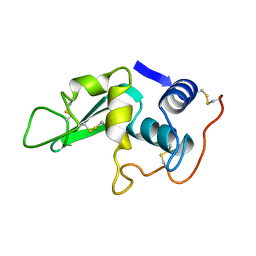 | | DISSECTION OF PROTEIN-CARBOHYDRATE INTERACTIONS IN MUTANT HEN EGG-WHITE LYSOZYME COMPLEXES AND THEIR HYDROLYTIC ACTIVITY | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Maenaka, K, Matsushima, M, Song, H, Watanabe, K, Kumagai, I. | | Deposit date: | 1995-02-10 | | Release date: | 1995-05-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissection of protein-carbohydrate interactions in mutant hen egg-white lysozyme complexes and their hydrolytic activity.
J.Mol.Biol., 247, 1995
|
|
3U6O
 
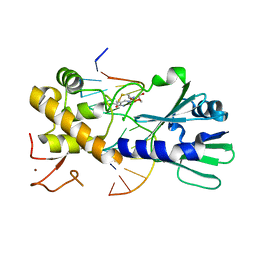 | | MutM set 1 ApG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*TP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*AP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
4EUH
 
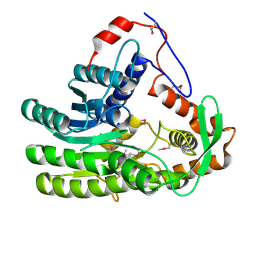 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase apo form | | Descriptor: | Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
2Y73
 
 | | THE NATIVE STRUCTURES OF SOLUBLE HUMAN PRIMARY AMINE OXIDASE AOC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Elovaara, H, Kidron, H, Parkash, V, Nymalm, Y, Bligt, E, Ollikka, P, Smith, D.J, Pihlavisto, M, Salmi, M, Jalkanen, S, Salminen, T.A. | | Deposit date: | 2011-01-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Two Imidazole Binding Sites and Key Residues for Substrate Specificity in Human Primary Amine Oxidase Aoc3.
Biochemistry, 50, 2011
|
|
4EPD
 
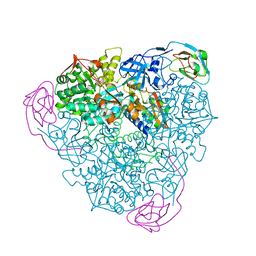 | | Initial Urease Structure for Radiation Damage Experiment at 300 K | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Warkentin, M, Badeau, R, Hopkins, J.B, Thorne, R.E. | | Deposit date: | 2012-04-17 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spatial distribution of radiation damage to crystalline proteins at 25-300 K.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EWN
 
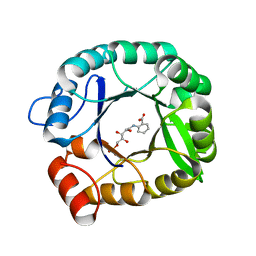 | | Structure of HisF-D130V+D176V with bound rCdRP | | Descriptor: | 1-(O-carboxy-phenylamino)-1-deoxy-D-ribulose-5-phosphate, Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Reisinger, B, Bocola, M, Rajendran, C, List, F, Sterner, R. | | Deposit date: | 2012-04-27 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | A sugar isomerization reaction established on various (beta-alpha)8-barrel scaffolds is based on substrate-assisted catalysis
Protein Eng.Des.Sel., 25, 2012
|
|
4EWW
 
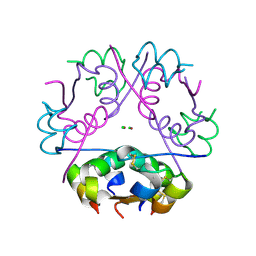 | | Human Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Favero-Retto, M.P, Palmieri, L.C, Lima, L.M.T.R. | | Deposit date: | 2012-04-28 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural meta-analysis of regular human insulin in pharmaceutical formulations.
Eur J Pharm Biopharm, 85, 2013
|
|
4EX0
 
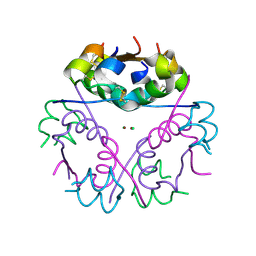 | | Human Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Favero-Retto, M.P, Palmieri, L.C, Lima, L.M.T.R. | | Deposit date: | 2012-04-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural meta-analysis of regular human insulin in pharmaceutical formulations.
Eur J Pharm Biopharm, 85, 2013
|
|
